Deep learning guided discovery of antibacterial polymeric nanoparticles
Deep learning guided discovery of antibacterial polymeric nanoparticles
Wu, Y.; Wang, C.; Shen, X.; Chen, Y.; Wang, H.; Xu, B.; Chen, Y.; Dai, W.; Huang, Y.; Zou, L.; Ji, J.; Zhang, P.
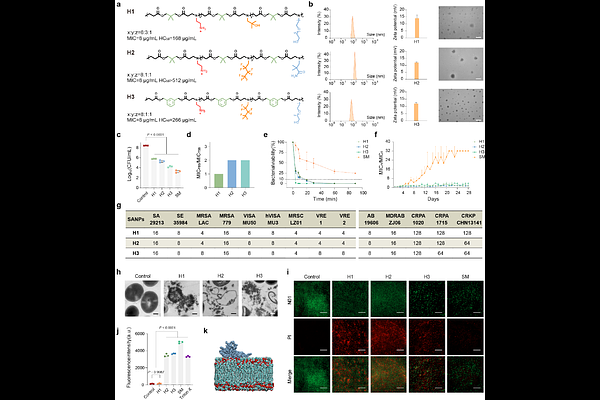
AbstractDrug-resistant bacterial infections pose a serious threat to global health, driving the development of novel antibacterial strategies beyond classic antibiotics. Self-assembled polymeric nanoparticles (SANPs) have emerged as promising candidates due to their unique structural properties, which enhance membrane interaction, drug loading, and therapeutic accumulation. The structural diversity and tunable properties of SANPs provide a flexible design platform for antibacterial applications. However, navigating the vast chemical space of SANPs remains a significant challenge due to the complex structure-activity relationships. To address this, we developed PolyCLOVER, a deep learning-guided framework for the efficient discovery of antibacterial SANPs. The framework integrates multi-stage self-supervised learning, active learning, and high-throughput experimentation to iteratively guide the discovery of SANPs with potent antibacterial activity and low toxicity. Through this framework, we identified three potent SANPs within a combinatorial library of ~100,000 poly({beta}-amino ester)s, with minimum inhibitory concentrations of 4 g/mL and 8 g/mL against clinically multidrug-resistant S. aureus and A. baumannii, respectively. Moreover, they can also act as antibiotic carriers, restoring the sensitivity of pathogens to penicillin G. In vivo studies demonstrated their therapeutic efficacy both as monotherapies and in combination therapies with antibiotics. These results demonstrate PolyCLOVER as a powerful framework for discovering multi-functional SANPs to combat antibiotic resistance.