MIC-Drop-seq: Scalable single-cell phenotyping of mutant vertebrate embryos
MIC-Drop-seq: Scalable single-cell phenotyping of mutant vertebrate embryos
Carey, C. M.; Parvez, S.; Brandt, Z. J.; Bisgrove, B. W.; Yates, C. J.; Peterson, R. T.; Gagnon, J. A.
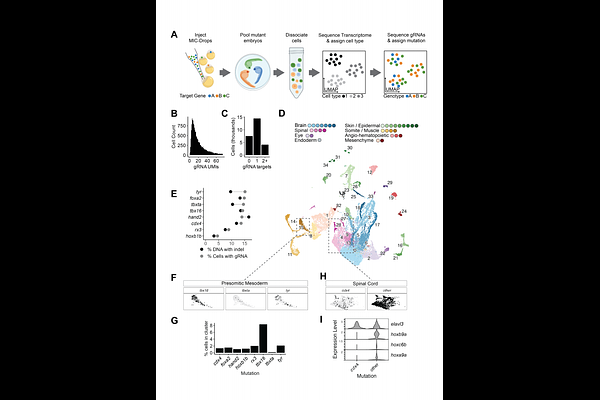
AbstractAdvances in genome engineering and single-cell RNA sequencing (scRNAseq) have revolutionized the ability to precisely map gene functions, yet scaling these techniques for large-scale genetic screens in animals remains challenging. We combined high-throughput gene disruption in zebrafish embryos via Multiplexed Intermixed CRISPR Droplets with phenotyping by multiplexed scRNAseq (MIC-Drop-seq). In one MIC-Drop-seq experiment, we intermixed and injected droplets targeting 50 transcriptional regulators into 1,000 zebrafish embryos, followed by pooled scRNAseq. Tissue-specific gene expression and cell abundance analysis of demultiplexed mutant cells recapitulated many known phenotypes, while also uncovering novel functions in brain and mesoderm development. We observed pervasive cell-extrinsic effects among these phenotypes, highlighting how whole-embryo sequencing captures complex developmental interactions. Thus, MIC-Drop-seq provides a powerful and scalable platform for mapping gene functions in vertebrate development with cellular resolution.