A genomic catalog of Earth's bacterial and archaeal symbionts
A genomic catalog of Earth's bacterial and archaeal symbionts
Villada, J. C.; Vasquez, Y.; Szabo, G.; Walker, E. W.; Gutierrez, M. R.; Qin, S.; Varghese, N.; Eloe-Fadrosh, E. A.; Kyrpides, N.; SymGs data consortium, ; Visel, A.; Woyke, T.; Schulz, F.
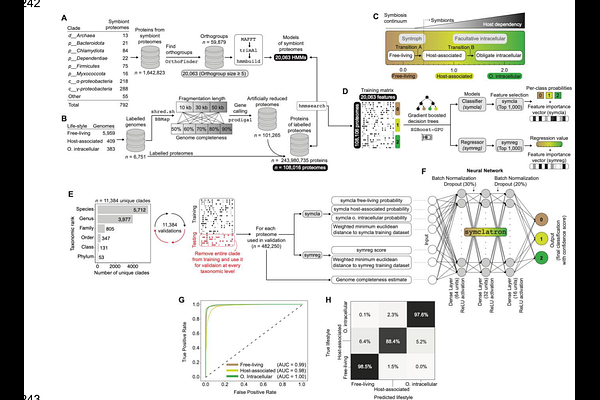
AbstractMicrobial symbiosis drives the functional and phylogenomic diversification of life on Earth, yet remains underexplored due to culturing challenges. This study employed machine learning (ML) to predict symbiotic lifestyles in hundreds of thousands of microbial genomes from diverse environmental metagenome samples and reference genomes. Predictions were performed using symclatron, a novel ML framework developed here to identify genomic signatures of symbionts. Predictions were deposited in a novel Symbiont Genomes catalog (SymGs). The results indicated that 15-23% of uncultivated microbes likely engage in symbiotic relationships with other organisms and are present in half of all known bacterial and archaeal phyla. We also identified genomic signatures of symbiotic lifestyles, including the loss of certain metabolic functions and the differential presence of others that may enable host-dependent living. The symclatron software and the SymGs catalog represent valuable resources for studying symbioses, facilitating future mechanistic investigations of host-microbe associations.