The effects of E. granulosus PSCs proteins on host immune cells were elucidated through Transcriptomic and Metabolomic analyses
The effects of E. granulosus PSCs proteins on host immune cells were elucidated through Transcriptomic and Metabolomic analyses
Aimulajiang, K.; Chen, X.; Aizezi, M.; Zhou, T.; Lin, R.; Wen, H.
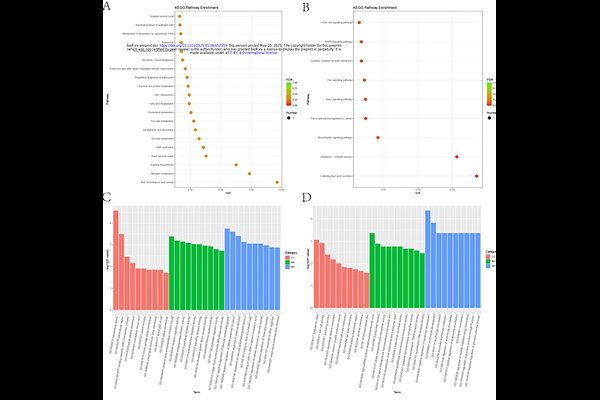
AbstractBackground Echinococcus granulosus (E. granulosus), a causative agent of zoonotic diseases, is widely prevalent across most regions and primarily affects vital organs such as the liver and lungs. The parasite secretes immunomodulatory substances, and its protoscolex proteins (PSCs) have been identified as potential diagnostic antigens and vaccine candidates. Although transcriptomics and metabolomics approaches have been employed to investigate echinococcosis, the impact of PSCs on the functionality of host immune cells and the expression of immunomodulatory-related genes remains largely unexplored. Methods In this study, immunofluorescence analysis was employed to determine the binding of PSCs to lymphocytes. The CCK-8 assay was utilized to evaluate the impact of varying concentrations of PSCs on cell viability, while ELISA was performed to quantify the levels of cytokines such as IL-9 and IL-4. Furthermore, by integrating LC-MS/MS-based metabolomics and RNA-Seq-based transcriptomics technologies, a systematic analysis was conducted to investigate the metabolic reprogramming and differential gene expression in lymphocytes following PSCs treatment, thereby elucidating the molecular mechanisms underlying its immune regulatory effects. Results PSCs proteins inhibited lymphocyte proliferation in a concentration-dependent manner, promoted IL-9 secretion at low concentrations, enhanced IL-4 expression at medium concentrations, and continuously up-regulated TGF-{beta} with increasing concentrations. Transcriptomic analysis identified 1,840 differentially expressed genes (991 up-regulated and 849 down-regulated), and KEGG enrichment analysis highlighted metabolic pathways, including phenylalanine metabolism. Gene Ontology (GO) analysis revealed that the functions of these genes were primarily associated with immunometabolic reprogramming and regulation of the intestinal barrier. Metabonomic profiling detected 12 significantly altered metabolites (8 up-regulated and 4 down-regulated). These findings suggest that PSCs reshape the host immune microenvironment via a metabolic-gene network. This study systematically elucidates how PSCs regulate immune cell function and metabolic homeostasis through multiple targets, providing novel insights into the immune escape mechanisms of parasites. Conclusion PSCs can induce host immune metabolic reprogramming, characterized by the activation of pyrimidine metabolism and the inhibition of bilirubin-mediated oxidative stress, through concentration-dependent regulation of Th9 cytokines (IL-9, IL-4/TGF-{beta}) and suppression of the PI3K-Akt signaling pathway. This process remodels the immune tolerance microenvironment. The aim is to elucidate the molecular mechanisms underlying parasite escape via multi-dimensional immunometabolic regulation, thereby providing a novel direction for the targeted treatment of echinococcosis.