Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Dereli, I.; Telychko, V.; Papanikos, F.; Raveendran, K.; Xu, J.; Boekhout, M.; Stanzione, M.; Neuditschko, B.; Imjeti, N. S.; Selezneva, E.; Erbasi, H. T.; Demir, S.; Giannattasio, T.; Gentzel, M.; Stevense, M.; Barchi, M.; Bondarieva, A.; Schnittger, A.; Weir, J.; Herzog, F.; Keeney, S.; Toth, A.
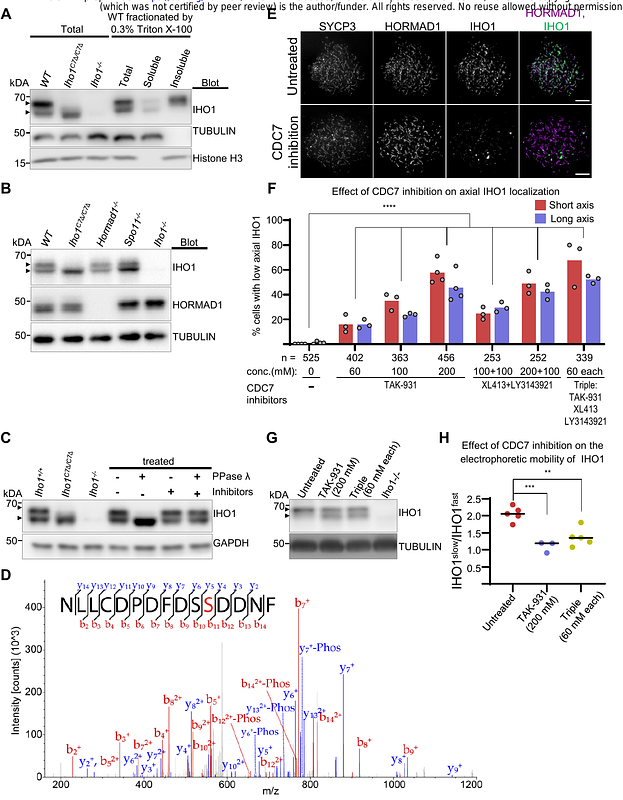
AbstractProgrammed DNA double-strand break (DSB) formation is a unique meiotic feature that initiates recombination-mediated linking of homologous chromosomes, thereby enabling chromosome number halving in meiosis. DSBs are generated on chromosome axes by heterooligomeric focal clusters of DSB-factors. Whereas DNA-driven protein condensation is thought to assemble the DSB-machinery, its targeting to chromosome axes is poorly understood. We discovered in mice that efficient biogenesis of DSB-machinery clusters requires seeding by axial IHO1 platforms, which are based on a DBF4-dependent kinase (DDK)-modulated interaction between IHO1 and the chromosomal axis component HORMAD1. IHO1-HORMAD1-mediated seeding of the DSB-machinery on axes ensures sufficiency of DSBs for efficient pairing of homologous chromosomes. Without IHO1-HORMAD1 interaction, residual DSBs depend on ANKRD31, which enhances both the seeding and the growth of DSB-machinery clusters. Thus, recombination initiation is ensured by complementary pathways that differentially support seeding and growth of DSB-machinery clusters, thereby synergistically enabling DSB-machinery condensation on chromosomal axes.