The Chlamydomonas reinhardtii CLiP2 mutant collection expands genome coverage with high-confidence disrupting alleles
The Chlamydomonas reinhardtii CLiP2 mutant collection expands genome coverage with high-confidence disrupting alleles
Lunardon, A.; Patena, W.; Pacini, C.; Warren-Williams, M.; Zubak, Y.; Laudon, M.; Silflow, C.; Lefebvre, P.; Jonikas, M. C.
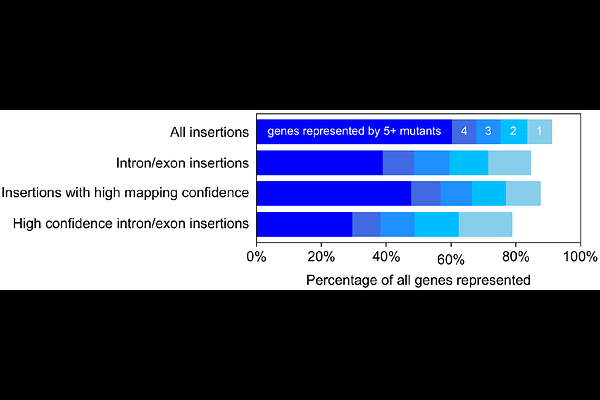
AbstractChlamydomonas reinhardtii (Chlamydomonas hereafter) is a powerful model organism for studies of photosynthesis, ciliary motility, and other cellular processes [1-4]. The CLiP library of mapped nuclear random insertion mutants [5,6] has accelerated progress for hundreds of laboratories in these fields by providing mutants in genes of interest. However, its value was limited by its modest coverage of the genome with high-confidence disruption alleles (46% of nuclear protein-coding genes with 1+ high-confidence allele in exons/introns; 12% of genes with 3+ alleles in exons/introns). Here we introduce the CLiP2 (Chlamydomonas Library Project 2) library, which greatly expands the number of available mapped high-confidence insertional mutants. The CLiP2 library includes 71,700 strains, covering 79% of nuclear protein-coding genes with 1+ high-confidence allele in exons/introns and 49% of genes with 3+ alleles in exons/introns. The mutants are available to the community via the Chlamydomonas Resource Center.