Force and the α-C-terminal domains bias RNApolymerase recycling
Force and the α-C-terminal domains bias RNApolymerase recycling
Qian, J.; Wang, B.; Artsimovitch, I.; Dunlap, D.; Finzi, L.
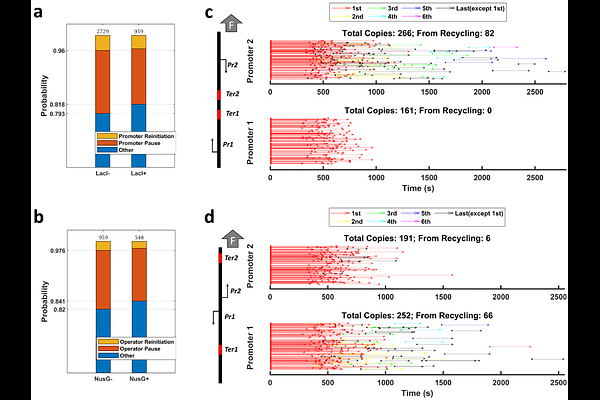
AbstractAfter RNA polymerase (RNAP) reaches a terminator, instead of dissociating from the template, it may diffuse along the DNA before restarting RNA synthesis from the previous or a different promoter. We monitored such secondary transcription using magnetic tweezers to determine the effect of low forces, protein roadblocks, and transcription factors. Up to 60% of RNAPs diffused along the DNA after termination. Force biased the direction of diffusion (sliding) and the velocity increased rapidly with force up to 0.7 pN and much more slowly thereafter. Sigma factor 70 ({sigma}70) likely remained bound to these sliding RNAPs, enabling recognition of secondary promoters and additional rounds of transcription, and the addition of elongation factor NusG, which competes with {sigma}70 for binding to RNAP, limited additional rounds of transcription. Surprisingly, RNAP blocked by a DNA-bound lac repressor could slowly re-initiate transcription at the blockage and was not affected by NusG, suggesting a {sigma}-independent pathway. Force biased the frequency of encounters between convergent or divergent promoters such that transcription was repetitive only from the promoter opposing the direction of force. Sliding RNAP recognized and could subsequently re-initiate from promoters in either orientation. However, deletions of the -C-terminal domains severely limited the ability of RNAP to turn around.