Conserved energetic changes drive function in an ancient protein fold
Conserved energetic changes drive function in an ancient protein fold
Wells, M. L.; Lu, C.; Sultanov, D.; Weber, K. C.; Gong, Z.; Glasgow, A.
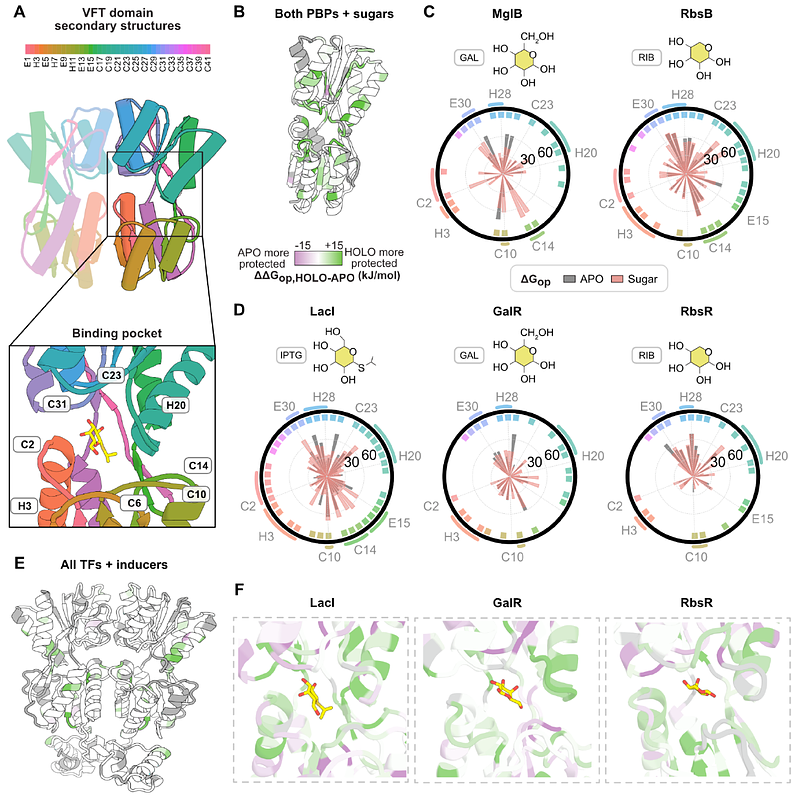
AbstractMany protein sequences occupy similar three-dimensional structures known as protein folds. In nature, protein folds are well-conserved over the course of evolution, such that there are 100,000 times as many extant protein sequences than there are folds. Despite their common shapes, similar protein folds can adopt wide-ranging functions, raising the question: are protein folds so strongly conserved for the purpose of maintaining function-driving energetic changes in protein families? Here we show that a set of key energetic relationships in a family of bacterial transcription factors (TFs) is conserved using high-resolution hydrogen exchange/mass spectrometry, bioinformatics, X-ray crystallography, and molecular dynamics simulations. We compared the TFs to their anciently diverged structural homologs, the periplasmic binding proteins (PBPs), expecting that protein families that share the same fold and bind the same sugars would have similar energetic responses. Surprisingly, our findings reveal the opposite: the \"energetic blueprints\" of the PBPs and the TFs are largely distinct, with the allosteric network of the TFs evolving specifically to support the functional requirements of genome regulation, versus conserved interactions with membrane transport machinery in PBPs. These results demonstrate how the same fold can be adapted for different sense/response functions via family-specific energetic requirements - even when responding to the same chemical trigger. Understanding the evolutionarily conserved energetic blueprint for a protein family provides a roadmap for designing functional proteins de novo, and will help us treat aberrant protein behavior in conserved domains in disease-related drug targets, where engineering selectivity is challenging.