Increasing mass spectrometry throughput using time-encoded sample multiplexing
Increasing mass spectrometry throughput using time-encoded sample multiplexing
Derks, J.; McDonnell, K.; Wamsley, N.; Stewart, P.; Yeh, M.; Specht, H.; Slavov, N.
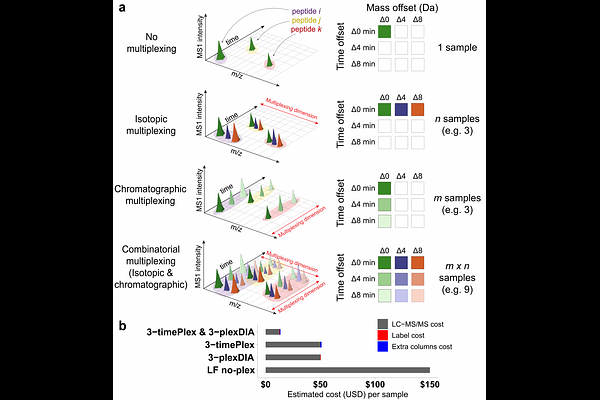
AbstractLiquid chromatography-mass spectrometry (LC-MS) can enable precise and accurate quantification of analytes at high-sensitivity, but the rate at which samples can be analyzed remains limiting. Throughput can be increased by multiplexing samples in the mass domain with plexDIA, yet multiplexing along one dimension will only linearly scale throughput with plex. To enable combinatorial-scaling of proteomics throughput we developed a complementary multiplexing strategy in the time domain, termed `timePlex\'. timePlex staggers and overlaps the separation periods of individual samples. This strategy is orthogonal to isotopic multiplexing, which enables combinatorial multiplexing in mass and time domains when paired together and thus multiplicatively increased throughput. We demonstrate this with 3-timePlex and 3-plexDIA, enabling the multiplexing of 9 samples per LC-MS run, and 3-timePlex and 9-plexDIA exceeding 500 samples / day with a combinatorial 27-plex. Crucially, timePlex supports sensitive analyses, including of single cells. These results establish timePlex as a methodology for label-free multiplexing and for combinatorially scaling the throughput of LC-MS proteomics. We project this combined approach will eventually enable an increase in throughput exceeding 1,000 samples / day.