Combining in-line chromatography coupled SAXS and AIpredicted structures to dissect the mechanism of ParB1parS1 partition assembly formation
Combining in-line chromatography coupled SAXS and AIpredicted structures to dissect the mechanism of ParB1parS1 partition assembly formation
ANU, A.; Lata, S.; Chaudhuri, B.
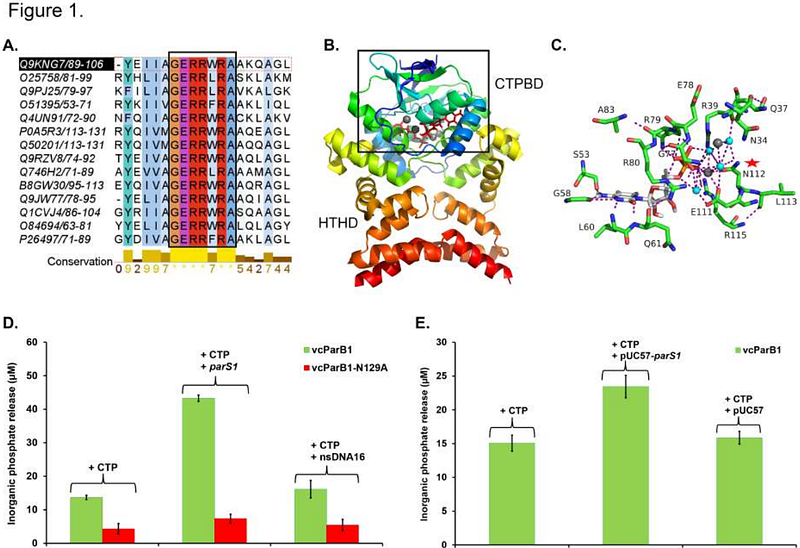
AbstractParB, which is a CTP-dependent DNA clamp, is an essential component of bacterial ParABS chromosomal origin segregation apparatus. A CTP-induced conformational switching leads to a closed conformation of dimeric ParB that displaces parS DNA from its binding site to enable sliding. We combined in-line chromatography-coupled SAXS and AI-predicted structure of full-length ParB1 from Vibrio cholerae to elucidate critical conformational changes associated with ParB1 parS1 assembly formation. Like other ParBs, ParB1 can hydrolyse CTP. In-line SAXS provided size distribution of ParB1, and showed population shift to a more compact state upon ParB1-parS1 assembling. A truncated N-terminal segment of ParB1 devoid of the putative intrinsically unstructured regions appears to self associate in multiple oligomeric states in solution. Imparting flexibility in a linker region joining this N-terminal segment and the C-terminal dimerization domain makes full-length ParB1 highly association-prone, implying that this linker may modulate self-interaction of vcParB1 for partition assembly condensate formation. To visualize the clamping-related conformational states, integrative models of full-length ParB1 and parS1-bound ParB1 were built by combining SAXS profiles with Alphafold2 models. Resultant integrative structural models revealed parS1-induced conformational changes in ParB1 at the onset of clamp formation.


