Shallow sequencing (Shall-seq) procedure: A method for determining the genetic lineages and detecting antimicrobial resistance genes of antimicrobial-resistant bacteria using minimum Nanopore sequencing data
Shallow sequencing (Shall-seq) procedure: A method for determining the genetic lineages and detecting antimicrobial resistance genes of antimicrobial-resistant bacteria using minimum Nanopore sequencing data
Yagi, N.; Miyagi, N.; Uechi, A.; Hirai, I.
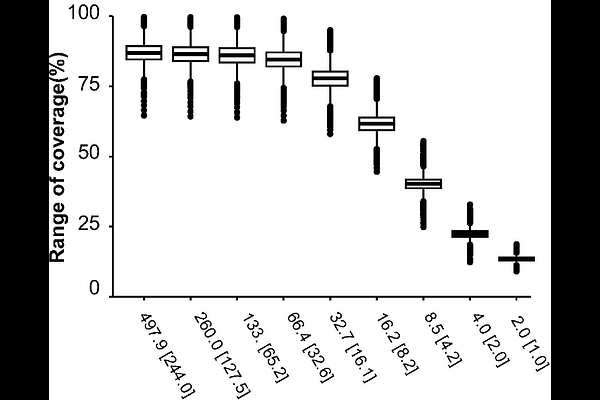
AbstractAntimicrobial-resistant bacteria could cause nosocomial infections and outbreaks in healthcare facilities. Phylogenetic analyses based on whole-genome sequencing (WGS) could become the gold-standard method for understanding the route of antimicrobial-resistant bacterial spreading. However, generally, the WGS needs to analyze much amount of data. Therefore, sufficient resources such as budget and data analysis system are needed and it is a burden for introduction of the WGS in the routine clinical examination of pathogenic bacterial isolates. In this study, we used Escherichia coli as a model and evaluated whether determination of the genetic background and detection of antimicrobial-resistance genes of 29 E. coli clinical isolates were achieved by searching databases using sequence reads output by the Nanopore sequencer as the search keys. Consequently, only 66.2 MB data was sufficient to search for a genome sequence with [≥]90% range of coverage rate. Importantly, AMR genes and plasmid replicon types were also detected with minimum data, and the detected AMR genes and phenotypes of the E. coli isolates did not present any discrepancy. Taken together, this shallow sequencing (Shall-seq) procedure consists of shallow of coverage sequencing using the Nanopore sequencer and data search using minimum data could be used to analyze bacterial isolates cost-effectively.