Genomic prediction of stalk lodging resistance and the associated intermediate phenotypes in maize using whole-genome resequence and multi-environmental data
Genomic prediction of stalk lodging resistance and the associated intermediate phenotypes in maize using whole-genome resequence and multi-environmental data
Machado e Silva, C.; Kunduru, B.; Bokros, N.; Tabaracci, K.; Oduntan, Y.; Brar, M. S.; Kumar, R.; Stubbs, C. J.; Nardino, M.; McMahan, C. S.; DeBolt, S.; Robertson, D. J.; Sekhon, R. S.; Morota, G.
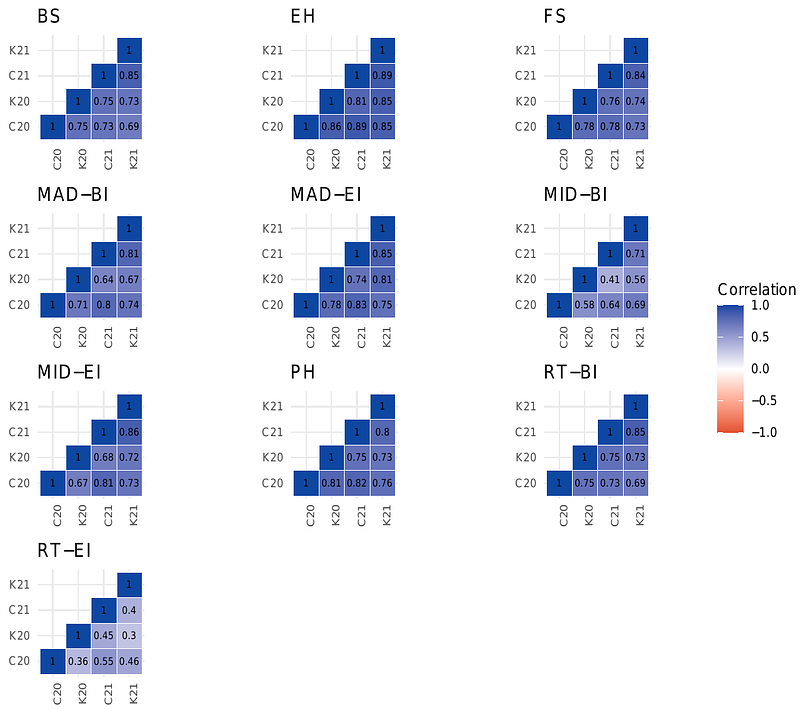
AbstractBreeding for stalk lodging resistance is of paramount importance to maintain and improve maize yield and quality and meet increasing food demand. The integration of environmental, phenotypic, and genotypic information offers the opportunity to develop genomic prediction strategies that can improve the genetic gain for complex traits such as stalk lodging. However, implementation of genomic predictions for stalk lodging resistance has been sparse primarily due to the lack of reliable and reproducible phenotyping strategies. In this study, we measured 10 traits related to stalk lodging resistance obtained from a novel phenotyping platform on approximately 31,000 individual stalks. These traits were combined with environmental information and whole-genome resequence data to investigate the predictive ability of different single and multi-environment genomic prediction models. In total, 555 maize inbred lines from the Wisconsin diversity panel were evaluated in four environments. The multi-environment models more than doubled the prediction accuracy compared to the single-environment model for most traits, particularly when predicting lines in a sparse testing design. Predictive correlations for stalk bending strength and stalk flexural stiffness, a non-destructive method for assessment of stalk lodging resistance, were moderately high and ranged between 0.32 to 0.89 and 0.26 to 0.88, respectively. In contrast, rind thickness was the most difficult trait to predict. Our results show that the use of multi-environmental data could improve genomic prediction accuracy for stalk lodging resistance and its intermediate phenotypes. This study will serve as a first step toward genetic improvement and the development of maize varieties resistant to stalk lodging.