Highly Repeatable Tissue Proteomics for Kidney Transplant Pathology: Technical and Biological Validation of Protein Analysis using LC-MS/MS
Highly Repeatable Tissue Proteomics for Kidney Transplant Pathology: Technical and Biological Validation of Protein Analysis using LC-MS/MS
Hofstraat, R.; Marx, K.; Blatnik, R.; Claessen, N.; Chojnacka, A.; Peters-Sengers, H.; Florquin, S.; Kers, J.; Corthals, G.
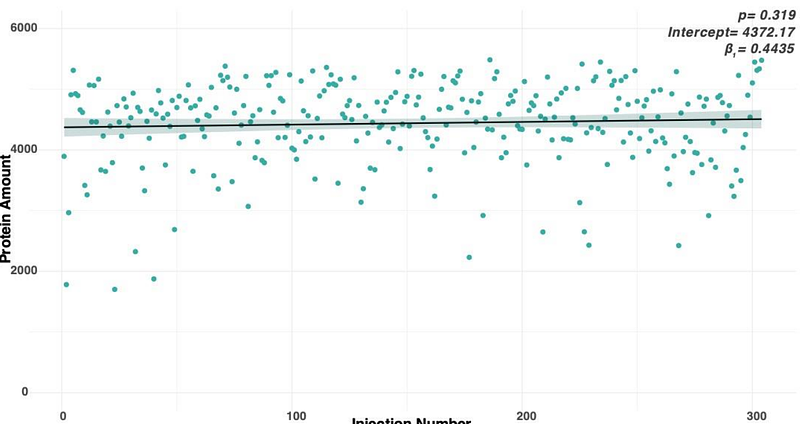
AbstractAccurate pathological assessment of tissue samples is key for diagnosis and optimal treatment decisions. Traditional pathology techniques suffer from subjectivity resulting in inter-observer variability, and limitations in identifying subtle molecular changes. Omics approaches provide both molecular evidence and unbiased classification, which increases the quality and reliability of final tissue assessment. Here, we focus on mass spectrometry (MS)-based proteomics as a method to reveal biopsy tissue differences. For MS data to be useful, molecular information collected from formalin fixed paraffin embedding (FFPE) biopsy tissues needs to be consistent and quantitatively accurate and contain sufficient clinically relevant molecular information. Therefore, we developed an MS-based workflow and assessed the analytical repeatability on 36 kidney biopsies, ultimately analysing molecular differences and similarities of over 5000 proteins per biopsy. Additional 301 transplant biopsies were analysed to understand other physical parameters including effects of tissue size, standing time in autosampler, and the effect on clinical validation. MS data were acquired using Data-Independent Acquisition (DIA) which provides gigabytes of data per sample in the form of high proteome (and genome) representation, at exquisitely high quantitative accuracy. The FFPE-based method optimised here provides a coefficient of variation below 20%, analysing more than 5000 proteins per sample in parallel. We also observed that tissue thickness does affect the outcome of the data quality: 5 m sections show more variation in the same sample than 10 m sections. Notably, our data reveals an excellent agreement for the relative abundance of known protein biomarkers with kidney transplantation lesion scores used in clinical pathological diagnostics. The findings presented here demonstrate the ease, speed, and robustness of the MS-based method, where a wealth of molecular data from minute tissue sections can be used to assist and expand pathology, and possibly reduce the inter-observer variability.


