Identification of Dehydrogenase, Hydratase, and Aldolase Required for C17 Propionyl Residue Removal in Steroid Degradation by Comamonas testosteroni TA441: Structural and Functional Insights into the (ChsE1-ChsE2)2 and (ChsH1-ChsH2MaoC)2-(Ltp2-ChsH2DUF35)2 Complexes
Identification of Dehydrogenase, Hydratase, and Aldolase Required for C17 Propionyl Residue Removal in Steroid Degradation by Comamonas testosteroni TA441: Structural and Functional Insights into the (ChsE1-ChsE2)2 and (ChsH1-ChsH2MaoC)2-(Ltp2-ChsH2DUF35)2 Complexes
Horinouchi, M.
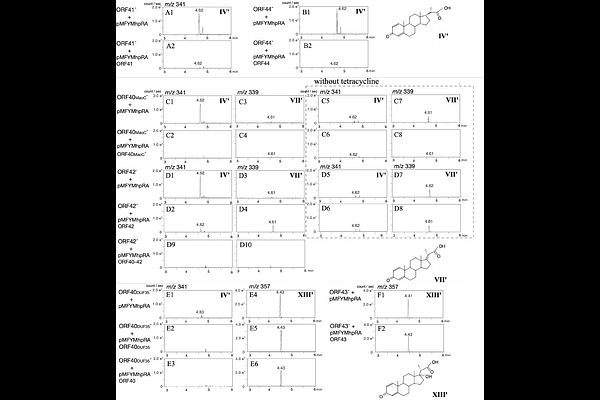
AbstractBacterial steroid degradation is gaining attention for its diverse roles, such as Mycobacteriumtuberculosiss reliance on the degradation of the C17 side chain of cholesterol for survival in host environments. ORF40 to ORF44 of Comamonastestosteroni TA441, previously characterized for its A-, B-, C-, and D-ring cleavage pathways, were hypothesized to correspond to the igr operon of M. tuberculosis, encoding the dehydrogenase ChsE1E2, hydratase ChsH1H2 maoC, and aldolase Ltp2 ChsH2DUF35, responsible for propionyl residue removal in C17 side-chain degradation. However, low amino acid identity between the corresponding enzymes precluded functional assignment without experimental evidence. In this study, we generated gene-disrupted mutants of ORF40-44 and demonstrated that ORF41/44, ORF40/42, and ORF43 encode the dehydrogenase, hydratase, and aldolase, respectively. ORF40 encodes a bifunctional protein comprising MaoC and DUF35 domains. The MaoC domain of ORF40 and the ORF42-encoded protein form the hydratase, while the DUF35 domain is essential for aldolase activity. A mutant expressing ORF40MaoC and ORF40DUF35 separately exhibited both hydratase and aldolase activities, suggesting these activities do not require a strictly formed complex of hydratase and aldolase. However, efficient propionyl residue removal appears to depend on the proper formation of each enzymatic complex, including ChsE1E2, ChsH1H2MaoC, and Ltp2ChsH2DUF35; although (ChsE1-ChsE2)2 does not form a stable complex with (ChsH1-ChsH2MaoC)2-(Ltp2-ChsH2DUF35)2, some degree of interaction was suggested. AlphaFold-predicted 3D structures of the TA441 enzymes and their complexes revealed striking similarities to those of M. tuberculosis, despite low amino acid identities. These findings shed light on the structural and functional conservation of bacterial steroid-degrading enzymes.