Hierarchical cross-entropy loss improves atlas-scale single-cell annotation models
Voice is AI-generated
Connected to paperThis paper is a preprint and has not been certified by peer review
Hierarchical cross-entropy loss improves atlas-scale single-cell annotation models
Cultrera di Montesano, S.; D'Ascenzo, D.; Raghavan, S.; Amini, A. P.; Winter, P.; Crawford, L.
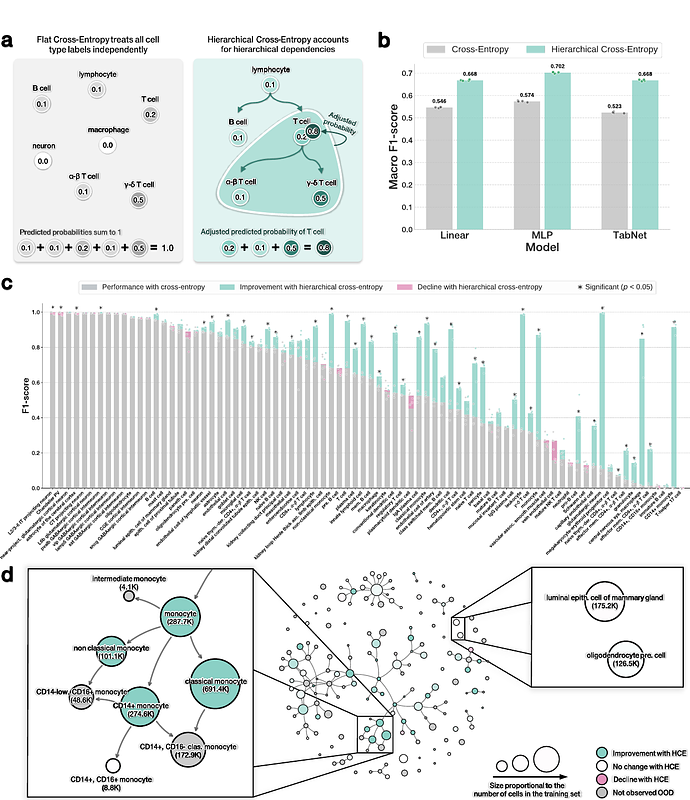
AbstractAccurately annotating cell types is essential for extracting biological insight from single-cell RNA-seq data. Although cell types are naturally organized into hierarchical ontologies, most computational models do not explicitly incorporate this structure into their training objectives. We introduce a hierarchical cross-entropy loss that aligns model objectives with biological structure. Applied to architectures ranging from linear models to transformers, this simple modification significantly improves out-of-distribution performance (12-15%) without added computational cost.