Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain
Allosteric modulation of the CXCR4:CXCL12 axis by targeting receptor nanoclustering via the TMV-TMVI domain
Garcia-Cuesta, E. M.; Martinez, P.; Selvaraju, K.; Gomez Pozo, A. M.; D'Agostino, G.; Gardeta, S.; Quijada-Freire, A.; Blanco Gabella, P.; Roca, C.; Jimenez-Saiz, R.; Garcia-Rubia, A.; Soler Palacios, B.; Lucas, P.; Ayala-Bueno, R.; Santander Acerete, N.; carrasco, y.; Martinez, A.; Campillo, N. E.; Jensen, L. D.; Rodriguez Frade, J. M.; Santiago, C.; Mellado, M.
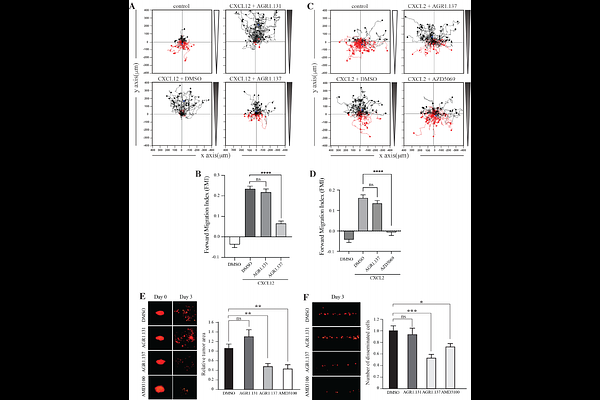
AbstractCXCR4 is a ubiquitously expressed chemokine receptor that regulates leukocyte trafficking and arrest in homeostatic and pathological states, and also participates in organogenesis, HIV-1 infection and tumor development. Despite the potential therapeutic benefit of CXCR4 antagonists, so far only one, plerixafor (AMD3100), which blocks the ligand-binding site, has reached the clinic. Recent advances in imaging and biophysical techniques have provided a richer understanding of the membrane organization and dynamics of this receptor. CXCL12 activation of CXCR4 reduces the number of CXCR4 monomers/dimers at the cell membrane and increases the formation of large nanoclusters, which are largely immobile and are required for correct cell orientation towards chemoattractant gradients. Mechanistically, CXCR4 activation involves a structural motif defined by residues on TMV and TMVI of CXCR4. Using this structural motif as a template, we performed in silico molecular modeling followed by in vitro screening of a small compound library to search for allosteric antagonists of CXCR4 that do not affect CXCL12 binding. We identified AGR1.137, a small compound that abolishes CXCL12-mediated receptor nanoclustering and dynamics and blocks the ability of cells to sense CXCL12 gradients both in vitro and in vivo without altering ligand binding or receptor internalization.