A Super-Loop Extrusion Mechanism Shapes the 3D Mitotic Chromosome Folding
A Super-Loop Extrusion Mechanism Shapes the 3D Mitotic Chromosome Folding
Norouzi, D.
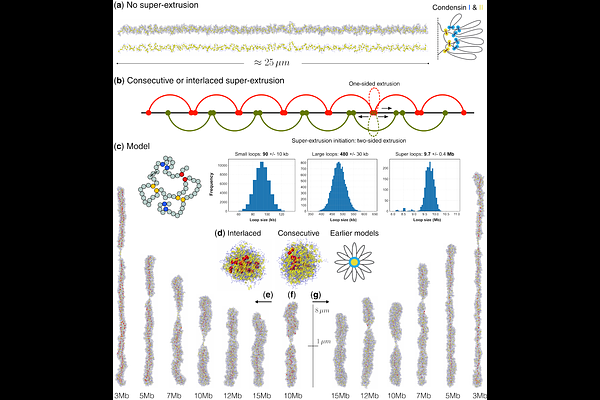
AbstractDespite significant advances in understanding mitotic chromosome folding, existing loop-extrusion models - based on two nested tiers of Condensin I- and II-mediated loops - fail to fully reproduce the structural features of chromosomes observed in vivo. We propose a hierarchical three-layer model in which a third set of megabase-scale \"super-loops,\" extruded by a small number of high-processivity motors, compacts chromatin to its native dimensions. Polymer simulations of human chromosome 1 (250 Mb) show that 25-200 consecutive or interlaced super-extruders forming ~10 Mb loops are sufficient to collapse a 25m bottle-brush structure into the empirically observed 5-8*1.5m rod. This model simultaneously reproduces the second diagonal in mitotic Hi-C maps, the double-helical scaffold seen in 3D-SIM microscopy, and the alternating-handedness, zero-mean helicity reported by high-resolution imaging. Interlaced super-loops enhance structural robustness against extruder loss and naturally account for the centromere as a region devoid of super-extruders. Although our simulations remain agnostic to molecular identity, we hypothesize that specialized forms of Condensin II, aided by auxiliary chromosome-associated proteins, act as super-extruders, while Topoisomerase II and other cross-linkers stabilize accumulated stress by converting transient DNA crossings into durable catenations. This three-layered mechanism resolves longstanding discrepancies in chromosome size, shape, contact-map profile, and scaffold ultrastructure within a unified physics-based framework. The next step is to identify or distinguish super-extruders experimentally and to design Hi-C, imaging, or mechanical perturbation assays to validate their existence and dynamics.