Leveraging a large language model to predict protein phase transition: a physical, multiscale and interpretable approach
Leveraging a large language model to predict protein phase transition: a physical, multiscale and interpretable approach
Frank, M.; Ni, P.; Jensen, M.; Gerstein, M. B.
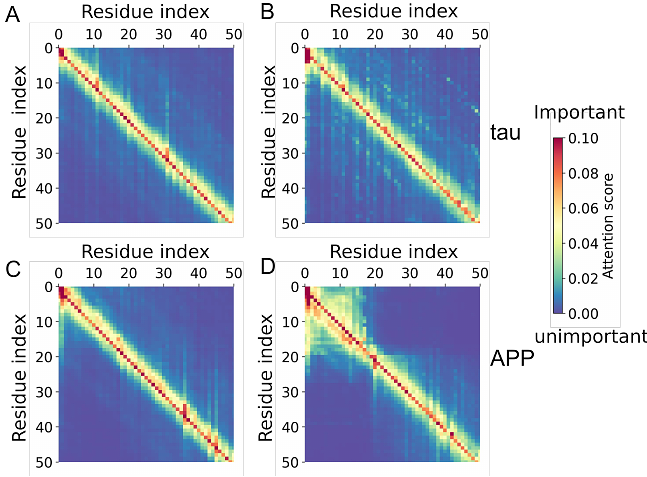
AbstractProtein phase transitions (PPTs) from the soluble state to a dense liquid phase (forming droplets via liquid-liquid phase separation) or solid aggregates (such as amyloid) play key roles in pathological processes associated with age-related diseases such as Alzheimer disease (AD). Several computational frameworks are capable of separately predicting the formation of protein droplets or amyloid aggregates based on protein sequences, yet none have tackled the prediction of both within a unified framework. Recently, large language models (LLMs) have exhibited great success in protein structure prediction; however, they have not yet been used for PPTs. Here, we fine-tune a LLM for predicting PPTs and demonstrate its superior performance compared to suitable classical benchmarks. Due to the\"black-box\" nature of the LLM, we also employ a classical random forest model along with biophysical features to facilitate interpretation. Finally, focusing on AD-related proteins, we demonstrate that greater aggregation is associated with reduced gene expression in AD, suggesting a natural defense mechanism.