RNA-sequencing Strain-specific Genome Alignment Increases Differential Expression Findings in Comparison of C57BL/6J and DBA/2J Nucleus Accumbens
RNA-sequencing Strain-specific Genome Alignment Increases Differential Expression Findings in Comparison of C57BL/6J and DBA/2J Nucleus Accumbens
Gnatowski, E.; Zeliff, D.; Dozmorov, M. G.; Miles, M. F.
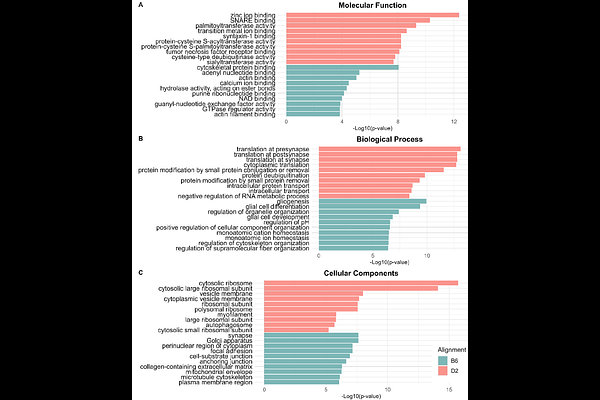
AbstractAlcohol Use Disorder (AUD) is a polygenic disease defined by the inability to regulate alcohol consumption despite adverse consequences. C57BL/6J (B6) and DBA/2J (D2) mice, the progenitor strains to the BXD recombinant inbred strain, exhibit differences in voluntary ethanol consumption and other ethanol behaviors, making them frequently used models for studying genetic influences on ethanol responses. The B6 genome is the standard reference genome for the majority of mouse RNA-sequencing (RNAseq) studies, including studies on D2 mice. We hypothesized that aligning B6 and D2 RNAseq data to their strain specific genome would allow improved detection of differentially expressed genes (DEGs) in comparison of brain gene expression between these two strains. RNA samples obtained from B6 and D2 nucleus accumbens (NAc) tissue were analyzed using a standard RNAseq analysis pipeline except from genome alignment. Following quality control, samples were aligned to either the B6 reference genome (Release 108) or the D2 samples were aligned to a recent homologous genome assembly (GCA_921998315.2). Alignment of D2 samples to the D2 genome showed significantly higher alignment compared to the B6 reference genome (93.82% vs 92.02%, p = 0.0272), but also showed a decrease in the number of total reads assigned (72.30%% vs 74.36%, p <0.0001). When comparing B6 and D2 expression, using the D2 alignment resulted in large increases in the number of differentially expressed genes (DEGs) (10,777 vs 6,191) and differentially utilized exons (DUEs) (81,206 vs 21,223) with resulting changes in gene ontology functional analyses. The gene ontology identified substantial overlap between the two analyses while also adding novel categories. These studies highlight the importance of using strain-specific alignment in increasing the number of reads aligned and the number of DEGs and DUEs identified. The use of strain-specific alignment in RNA-seq studies may provide greater accuracy in investigating gene expression and pathways regulated by ethanol in model organism studies on molecular mechanisms of AUD.