Targeted loss of heterozygosity in Candida albicans using CRISPR-Cas9
Targeted loss of heterozygosity in Candida albicans using CRISPR-Cas9
Despres, P. C.; Gervais, N. C.; Fogal, M.; Rogers, R. K.; Cuomo, C. A.; Shapiro, R. S.
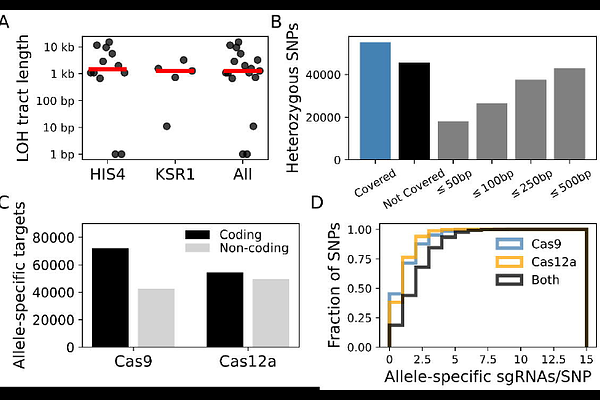
AbstractThe diploid genome of the fungal pathogen Candida albicans is highly heterozygous, with most allele pairs diverging at either the coding or regulatory level. When faced with selection pressure like antifungal exposure, this hidden genetic diversity can provide a reservoir of adaptive mutations through loss of heterozygosity (LOH) events. Validating the potential phenotypic impact of LOH events observed in clinical or experimentally evolved strains can be difficult due to the challenge of precisely targeting one allele over the other. Here, we show that a CRISPR-Cas9 system can be used to overcome this challenge. By designing allele-specific guide RNA sequences, we can induce targeted, directed LOH events, which we validate by whole-genome long-read sequencing. Using this approach, we efficiently recapitulate a recently described LOH event that increases resistance to the antifungal fluconazole. Additionally, we find that the recombination tracts of these induced LOH events have similar lengths to those observed naturally. To facilitate future use of this method, we provide a database of allele-specific sgRNA sequences for Cas9 that provide near genome-wide coverage of heterozygous sites through either direct or indirect targeting. Finally, we provide tools to generate custom databases for any diploid species or targeted nuclease of interest. This approach will be useful in probing the adaptive role of LOH events in this important human pathogen.