An Archaeal Transcription Factor Bridges Prokaryotic and Eukaryotic Regulatory Paradigms
An Archaeal Transcription Factor Bridges Prokaryotic and Eukaryotic Regulatory Paradigms
Ferrer, F. M.; Nayak, D. D.
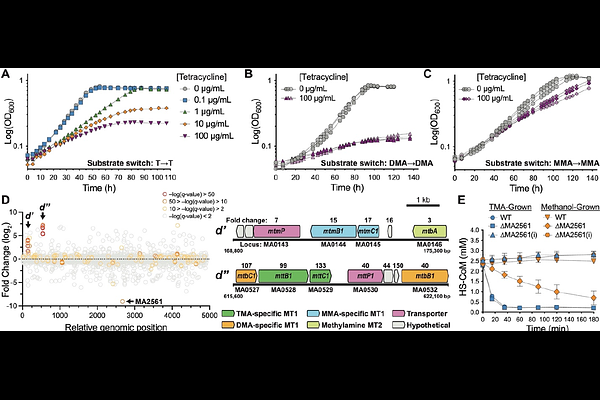
AbstractArchaeal transcription is a hybrid of eukaryotic and bacterial features: a RNAP II-like polymerase transcribes genes organized in circular chromosomes within cells devoid of a nucleus. Consequently, archaeal genomes are depleted in canonical transcription factors (TFs) found in other domains of life. Here we outline the discovery of a cryptic archaea-specific family of ligand-binding TFs, called AmzR (Archaeal Metabolite-sensing Zipper-like Regulators). We identify AmzR through an evolution-based genetic screen and show that it is a negative regulator of methanogenic growth on methylamines in the model archaeon, Methanosarcina acetivorans. AmzR binds its target promoters as an oligomer using paired basic -helices akin to eukaryotic leucine zippers. AmzR also binds methylamines, which reduces its DNA-binding affinity, and allows it to function as a one-component system commonly found in prokaryotes, while containing a eukaryotic-like DNA-binding motif. The AmzR family of TFs are widespread in archaea and broadens the scope of innovations at the prokaryote-eukaryote interface.