Convergent evolution in toxin detection and resistance provides evidence for conserved bacterial-fungal interactions
Convergent evolution in toxin detection and resistance provides evidence for conserved bacterial-fungal interactions
Dolan, S. K.; Duong, A. T.; Whiteley, M.
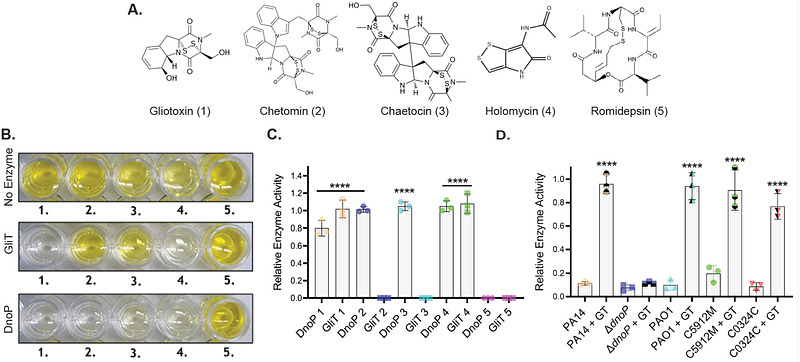
AbstractMicrobes rarely exist in isolation, and instead form complex polymicrobial communities. As a result, microbes have developed intricate offensive and defensive strategies that enhance their fitness in these complex communities. Thus, identifying and understanding the molecular mechanisms controlling polymicrobial interactions is critical for understanding the function of microbial communities. In this study, we show that the Gram-negative opportunistic human pathogen Pseudomonas aeruginosa, which frequently causes infection alongside a plethora of other microbes including fungi, encodes a genetic network which can detect, and defend against gliotoxin, a potent, disulfide-containing antimicrobial produced by the ubiquitous filamentous fungus Aspergillus fumigatus. We show that gliotoxin exposure disrupts P. aeruginosa zinc homeostasis, leading to transcriptional activation of a gene encoding a previously uncharacterized dithiol oxidase (DnoP), which detoxifies gliotoxin and structurally related toxins. While the enzymatic activity of DnoP is identical to that used by A. fumigatus to protect itself against gliotoxin, DnoP shares little homology to the A. fumigatus gliotoxin resistance protein. Thus, DnoP and its transcriptional induction by low zinc represent an example of both convergent evolution of toxin defense and environmental cue sensing across kingdoms. Collectively, these data support disulfide-containing natural products as mediators of inter-kingdom interactions and provide evidence that P. aeruginosa has evolved to survive exposure to these molecules in the natural environment.