A major update of the genome assembly of Eri silkmoth, Samia ricini.
A major update of the genome assembly of Eri silkmoth, Samia ricini.
Lee, J.; Okamoto, M.; Kawagoe, R.; Shimada, T.
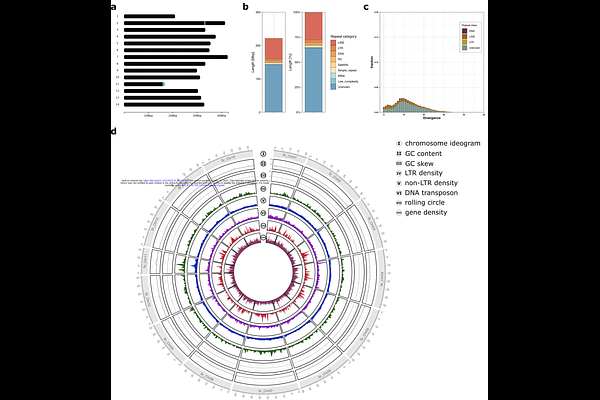
AbstractIndian eri silkmoth, Samia ricini, is a wild silkmoth whose silk occupies a significant economic position. In addition to its importance as an economic animal, S. ricini is also useful as a model species of Saturniidae. National BioResource of Japan (NBRP) maintains a S. ricini strain brought to Japan during WWII via Taiwan. Since we have previously published a draft genome assembly of S. ricini, we have attempted to construct a chromosome-level genome assembly to facilitate genetic studies of S. ricini. We successfully constructed a chromosome-scale genome assembly by exploiting two long-read-based technologies, HiFi reads and optical genome mapping. Furthermore, we performed functional annotations of the genome assembly, i.e., repeat annotation, transcriptome-based gene prediction, ATAC-seq, and PIWI-interacting RNA (piRNA)-targeted small RNA-seq. The assembly harbours 16,226 protein-coding genes and 636 piRNA clusters across three tissues: ovaries, testis, and embryos. ATAC-seq data comprehensively detected open chromosome regions, which will benefit when CRISPR/Cas9-mediated genome editing is conducted.