Adapter dilution and input optimisation for Qiagen QIAseq miRNA Library kit
Adapter dilution and input optimisation for Qiagen QIAseq miRNA Library kit
Hasby, F.; Bachmann, J. A.; Wang, C.; Contreras-Lopez, O.; Mezger, A.
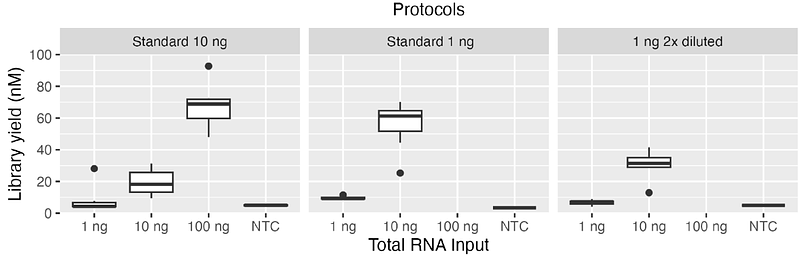
AbstractMicroRNAs (miRNAs) are small, non-coding RNA that play a critical role in regulating gene expressions that are important for a multitude of biological processes. In the preparation of miRNA sequencing libraries by using QIAseq miRNA library kit, adapter dimers might occur inadvertently and compromise the sequencing performance. In this technical note, we tested different adapter dilution strategies to minimise or diminish adapter dimers without compromising data quality. In our experiments, we found that RNA input amount plays a larger role in adapter dimer formation rather than the availability of the adapters itself. We suggest preparing the sequencing library with 10 times more RNA than the recommended input amount of the chosen protocol, e.g. following the adapter dilution and PCR cycle number of the standard 1 ng protocol with 10 ng RNA input. We also suggest analysing the data with deduplication by using unique molecular identifiers (UMIs) as it helps remove unusable reads.