Testing for a wildlife reservoir of divergent SARS-CoV-2 in white-tailed deer
Testing for a wildlife reservoir of divergent SARS-CoV-2 in white-tailed deer
Crawshaw, L.; Kotwa, J. D.; Jeeves, S. P.; Loomer, C.; Dibernardo, A.; Stewart, A.; Newar, S. L.; Chien, E.; Yim, W.; Kruczkiewicz, P.; Vernygora, O.; Lung, O.; Schulte-Hostedde, A. I.; Maguire, F.; Pickering, B.; Jardine, C. M.; Coatsworth, H.; Mubareka, S.; Bowman, J.
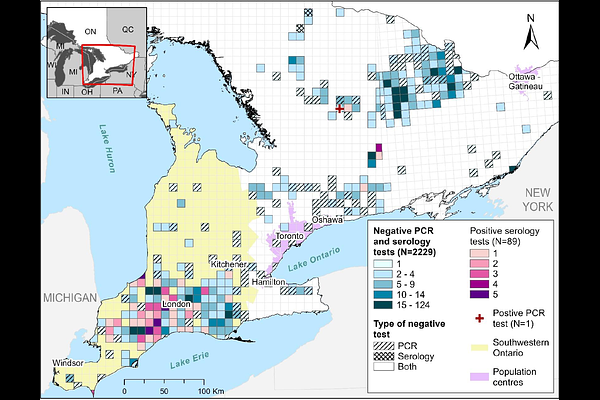
AbstractThe 2021 discovery of a divergent lineage (B.1.641) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in white-tailed deer (Odocoileus virginianus) from Ontario raised concerns that deer were a potential reservoir. To assess whether white-tailed deer continued to be infected with B.1.641 and to test for spillover into other species, we established a surveillance program in Ontario by sampling wildlife via existing monitoring programs and through active surveillance of captive and wild animals. Between 2022 to 2024, we tested 2,839 animals, identifying one active SARS-CoV-2 infection (a likely spillover of a recombinant XBB.2.3.11.3 lineage), but no cases of B.1.641. Overall, 93 animals (6.8%) tested positive for SARS-CoV-2 antibodies, including 89 white-tailed deer, two Virginia opossums (Didelphis virginiana), one American mink (Neogale vison), and one river otter (Lontra canadensis). In Southwestern Ontario, where B.1.641 was originally detected, 15.2% of deer samples were seropositive. Generalized Linear Models demonstrated that seropositive deer were more likely to be found in areas with a higher fall deer harvest and human population density, and closer to previous B.1.641 cases. Our data suggest that deer-associated B.1.641 may have caused a relatively localized epizootic without forming a stable reservoir. This study underscores the importance of One Health-focused surveillance.