Helfrich Monte Carlo Flexible Fitting: physics-based, data-driven cell-scale simulations
Helfrich Monte Carlo Flexible Fitting: physics-based, data-driven cell-scale simulations
Maurer, V. J.; Siggel, M.; Jensen, R. K.; Mahamid, J.; Kosinski, J.; Pezeshkian, W.
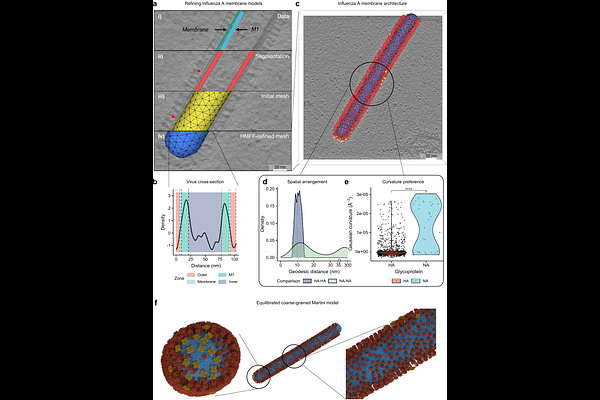
AbstractSimulating entire cells represents the next frontier of computational biology. Achieving this goal requires methods that accurately reconstruct and simulate cellular membranes across spatial and temporal scales. Although three-dimensional electron microscopy enables detailed membrane visualization, dose limitations and restricted field-of-view often result in fragmented membrane representations incompatible with simulations. To resolve this, here we introduce Helfrich Monte Carlo Flexible Fitting (HMFF), a novel approach that integrates experimental density data into membrane simulations. Together with the accompanying Mosaic software ecosystem, HMFF translates static imaging data into complete and physically regularized simulation-ready models at micrometer scales. We demonstrate the versatility of HMFF across diverse biological systems, including influenza virus particles, Mycoplasma pneumoniae cells, and entire eukaryotic organelles. These models enable multi-scale simulations involving millions of lipids, incorporate proteins at experimentally determined positions, and allow for quantitative morphological analysis of cellular compartments. HMFF thus establishes a foundation for data-driven whole-cell simulations.