Personalized Gut-Liver Microphysiological System Maps Donor-Specific Tissue Resident Immunity and Reveals a Conserved Metabolic Crosstalk
Personalized Gut-Liver Microphysiological System Maps Donor-Specific Tissue Resident Immunity and Reveals a Conserved Metabolic Crosstalk
Uslu, M.; Ran, R.; Siddiqui, M. F.; Liang, J.; Raechal, L.; Dogsa, M.; Perrot, C.; Lieberman, L. A.; Brubaker, D. K.; Trapecar, M.
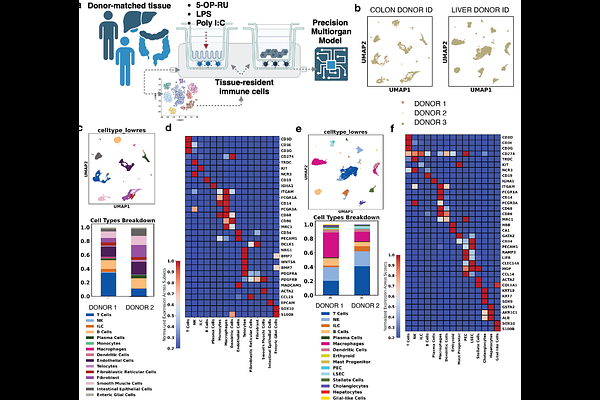
AbstractTissue-resident immune (TRI) niches underpin barrier defense yet differ markedly between individuals, complicating efforts to model inter-organ immunity in vitro. Here we combine single-cell atlases with engineered biology to build a personalized, immunocompetent gut-liver microphysiological system (MPS). Single-cell RNAseq of paired colon and liver from healthy donors reveals pronounced donor-specific TRI composition: Donor 1 is enriched for tissue-resident memory T cells and inflammatory macrophages, whereas Donor 2 harbors abundant plasma cells, MAIT cells. We re-create two donors in vitro by integrating primary epithelium, hepatocytes and their autologous CD45+ TRI populations into a low-shear, 3D-printed MOTIVE-2 vessel that fluidically couples gut and liver compartments. Local co-culture establishes parenchyma-specific immune programs: colonic epithelia drive Th1/Th17 skewing in Donor 1 but B-cell differentiation in Donor 2. Strikingly, once organs share medium under flow, both donors converge on a shared retinoid-bile acid-metabolic axis and a muted inflammatory set-point, demonstrating that systemic metabolite exchange can override baseline immune divergence. Intestinal challenge with microbial agonists uncovers stimulus- and donor-specific early liver responses: Poly I:C elicits a uniform type-I/III interferon burst, but LPS provokes stronger reactions in macrophage-rich Donor 1, whereas the riboflavin metabolite 5-OP-RU selectively activates MAIT-enriched Donor 2. These functional outputs align with ligand-receptor predictions and TRI composition, validating the platform's capacity to forecast personalized immunodynamics. Together, our study delivers a donor-resolved, immune-competent gut-liver MPS and reveals how conserved metabolic dialogue co-exists with, and is modulated by, individual TRI fingerprints. The approach enables "clinical-trial-on-chip" exploration of immunometabolic diseases and precision therapeutics, providing a blueprint for next-generation multiorgan models that honor human immune diversity.