eLaRodON: identification of large genomic rearrangements in Oxford Nanopore sequencing data
eLaRodON: identification of large genomic rearrangements in Oxford Nanopore sequencing data
Mikheeva, R.; Koryukov, M.; Leonov, P.; Mukhametdinova, V.; Borobova, V.; Laktionov, P.; Tarkhov, A.; Filipenko, M.; Kechin, A.
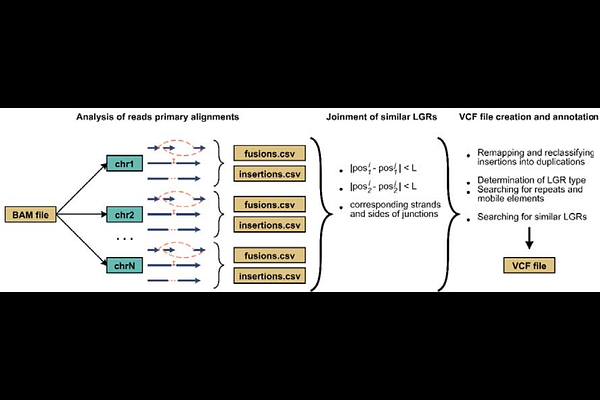
AbstractLong-read sequencing enables more accurate detection of large genomic rearrangements (LGRs) compared to short-read technologies. However, existing tools for LGR calling continue to evolve and require further optimization. In this study, we present eLaRodON, a novel tool for identifying LGRs in Oxford Nanopore sequencing data. Using publicly available datasets--including Mycobacterium tuberculosis genomes with extensive complete genome references and the human cell line NA12878--we demonstrate that eLaRodON outperforms existing tools (NanoSV, Sniffles2, NanoVar, and SVIM), achieving an AUC of 0.61 (vs. 0.13-0.43) for M. tuberculosis and 0.86 (vs. 0.40-0.72) for the human genome. Validation against gold-standard LGR sets (derived from de novo Flye assemblies and Mauve alignments) confirmed the tool\'s high accuracy. Notably, we identified recurrent false-positive LGR patterns across diverse datasets (M. tuberculosis, NA12878, and {lambda} phage controls). Orthogonal validation by targeted NGS and Sanger sequencing yielded a variant verification rate of 67-100% for different LGR types--including those supported by a single read. These results represent a significant advancement in LGR detection accuracy, with implications for genomic research and clinical applications. eLaRodON is an open-source program, and its code can be freely accessed on GitHub: https://github.com/aakechin/eLaRodON/.