From lab to field: analyses of genome-edited bacterial blight resistant rice
From lab to field: analyses of genome-edited bacterial blight resistant rice
Frommer, W. B.; Loo, E. P.; Huguet-Tapia, J. C.; Selvaraj, M.; Stiebner, M.; Killing, B.; Buchholzer, M.; Schepler-Luu, V.; Hartwig, T.; Valdez Gutierrez, S.; Rast-Somssich, M. I.; Szurek, B.; Tohme, J.; Charraviaga, P.; White, F. F.; Yang, B.
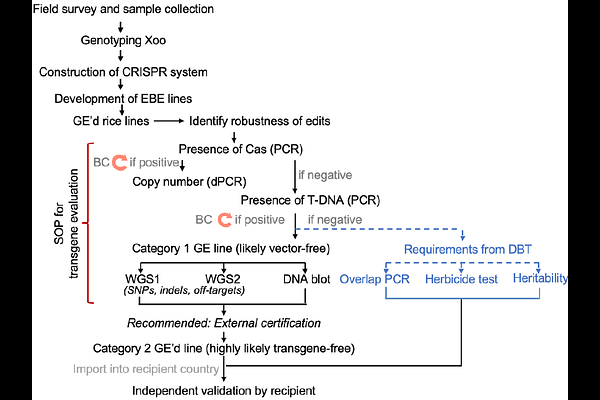
AbstractBacterial blight (BB) of rice, caused by Xanthomonas oryzae pv. oryzae (Xoo) is one of the major drivers of yield losses in Africa and Asia. Xoo secretes TAL-effectors (TALe) that induce host SWEET sucrose uniporter by binding to the effector binding element (EBE) of SWEET promoters, likely required for Xoo reproduction and virulence. We had multiplex edited the EBEs of three SWEET genes to prevent TALe binding, producing genome-edited (GE-d) rice mega-varieties (IR64, Ciherang-Sub1 for Asia, and Komboka for Africa) that were resistant to a wide spectrum of Xoo strains. Here, we report comprehensive analyses of the GE-d lines, including evaluation of agronomic performance in multi-location multi-season experimental field plots under different fertilization regimes, and tests for the presence/absence of foreign DNA/transgene in the offspring of GE-d lines (IR64-BC1T6, Ciherang-Sub1-BC1T5, Komboka-T3). Various strategies were evaluated, including herbicide tolerance, PCR, DNA gel blotting, whole genome sequencing (WGS), and specific tests stipulated by country-specific biosafety guidelines. Different WGS technologies were evaluated and also used to identify heritability of the edits, single nucleotide polymorphisms (SNPs), and insertions/deletions (indels) that might have resulted from somaclonal variation and potential GE-induced off-target mutations. Complete genome reference sequences for the parental lines IR64, Ciherang-Sub1, and Komboka are provided. In the field experiments, the GE-d lines did not show performance defects. Together, the results indicate that select GE-d lines do not contain foreign DNA or transgene fragments and fulfill the requirements for treatment equivalent to classical breeding lines in countries such as India and Kenya.