CellCousin2: An Optimized System for Partial Ablation and Tracing of Regenerative Lineages
CellCousin2: An Optimized System for Partial Ablation and Tracing of Regenerative Lineages
Hovhannisyan, G. G.; Akhourbi, T.; Eski, S. E.; Pirson, I.; Gurzov, E. N.; Singh, S. P.
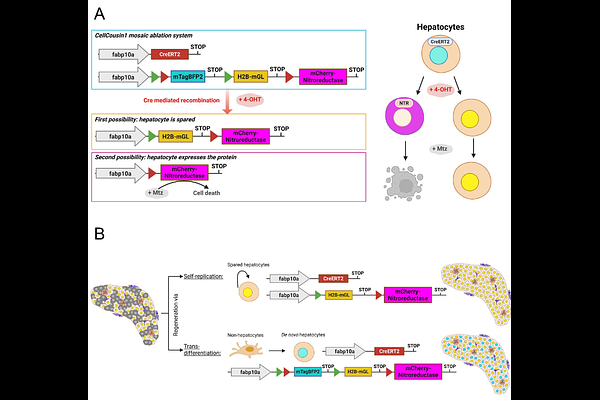
AbstractRegeneration can rely on multiple cellular sources, including stem cells, self-duplicating cells, and transdifferentiating cells. A central question in regenerative biology is how these distinct lineages contribute to repair and interact within a functional regenerate. We previously developed the CellCousin system to study cellular plasticity using inducible recombination and nitroreductase-mediated ablation in zebrafish. Here, we present CellCousin2, which introduces two key improvements for long-term tracking of spared and regenerating cells. First, to reduce background recombination, we developed a Dihydrofolate Reductase (DHFR)-CreER system with dual control: DHFR-mediated degradation in the absence of trimethoprim, and tamoxifen-dependent activation. This combination minimizes leakiness while maintaining high recombination efficiency. Second, we replaced the original nitroreductase with NTR2.0, enabling effective ablation with tenfold lower metronidazole concentration, reducing off-target effects on the liver. Together, these enhancements make CellCousin2 a robust platform for dissecting the dynamics and interactions of regenerative lineages.