Two high-quality rose genomes underpin a novel Rosa pangenome to advance rose genomics, phylogenetics, and breeding
Two high-quality rose genomes underpin a novel Rosa pangenome to advance rose genomics, phylogenetics, and breeding
Yang, Z.; Schijlen, E. G. W. M.; Schranz, E.; Zwaan, B. J.; de Ridder, D.; Visser, R. G. F.; Arens, P. F. P.; Smulders, M. J. M.; van Velzen, R.; Keurentjes, J. J. B.; Bourke, P. M.; Smit, S.
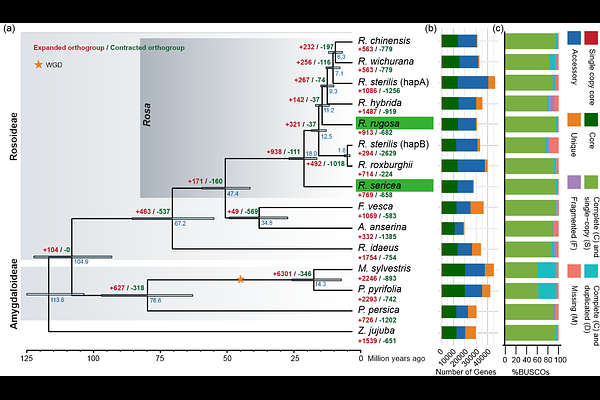
AbstractRosa, belonging to the family Rosaceae, encompasses more than 150 species which are widely distributed in the northern hemisphere. Renowned for their beauty, roses are cultivated throughout the world for ornamental purposes and the production of essential oils and perfumes. Despite their cultural and commercial significance, the genomic resources of wild Rosa species have not been studied comprehensively, hampering the understanding of their genetic diversity, evolutionary history, and breeding potential. Here we report on high-quality de novo genomes for Rosa sericea and Rosa rugosa. By integrating these two de novo genomes with existing public genomic resources, we have built a Rosaceae panproteome and a Rosa pangenome (spanning wild, traditional garden, and modern rose lineages) using a De Bruijn graph (DBG)-based approach. A maximum likelihood (ML) phylogeny of 18 Rosa haplotypes based on 4,367 single-copy core homology groups (genes) provided robust evolutionary inference, confirming the basal position of R. sericea, and enabled a gene-based macrosynteny analysis across the pangenome. Our analyses revealed significant genomic diversity among species, extensive variation in core gene content, and lineage-specific transposable element (TE) expansion patterns that contribute to the variation in Rosa genome size and to species-specific adaptations. The pangenome also revealed biased diversification of homology groups potentially linked to phenotypic plasticity in Rosa. Specifically, our analysis of the rose scent-related gene family, NUDX1, uncovered its evolutionary trajectory in Rosa, in which TEs insertions provided putative novel regulatory elements that facilitated adaptive evolution in metabolic pathways. This pangenomic study deepens our understanding of the genetic diversity and evolution of traits within the Rosa genus. In addition, the findings lay the foundation for future efforts to understand the genetic mechanisms driving trait evolution, which can support rose breeding.