Chromosome-level genome assembly of autotetraploid Selenicereus megalanthus and gaining genomic insights into the evolution of trait patterning in diploid and polyploid pitaya species
Chromosome-level genome assembly of autotetraploid Selenicereus megalanthus and gaining genomic insights into the evolution of trait patterning in diploid and polyploid pitaya species
Zaman, Q. U.; Hui, L.; Nazir, M. F.; Wang, G.; Garg, V.; Ikram, M.; Raza, A.; Lv, W.; Khan, D.; Khokhar, A. A.; You, Z.; Chitikineni, A.; Usman, B.; Jianpeng, C.; Yang, X.; Zuo, S.; Liu, P.; Kumar, S.; Guo, M.; Zhu, Z.-X.; Dwivedi, G.; Qin, Y.; Varshney, R. K.; Wang, H.-F.
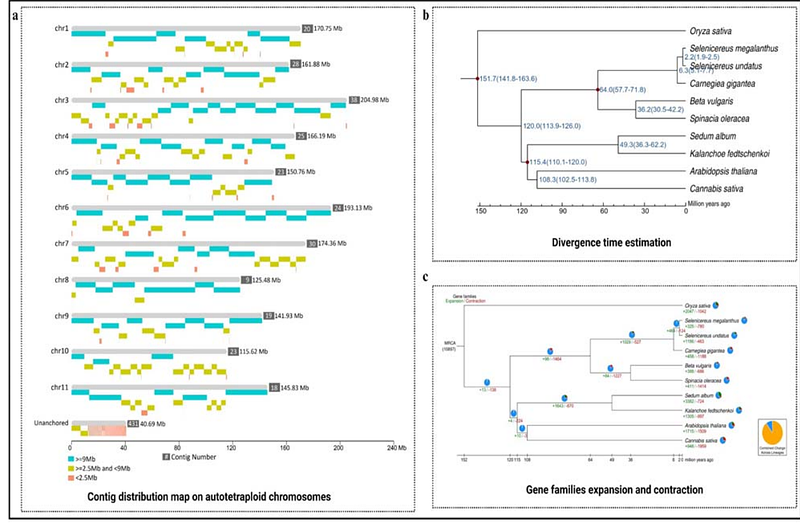
AbstractYellow pitaya (Selenicereus megalanthus, 2n=4x=44) breeding remains severely hindered due to lacking a reference genome. Here we report yellow pitaya\'s high-quality chromosome-level genome assembly and link the phenotypic trait with genomic data, based on Hi-C, ATAC, and RNA-seq data of specific tissues. We declared yellow pitaya as an autotetraploid with a 7.16 Gb genome size (harboring 27,246 high confidence genes) and majorly evolved from diploid ancestors, which remains unknown. Beyond generating the genome assembly, we explored the 3D chromatin organization which revealed insights into the genome, compartments A (648 and 519), compartments B (728 and 1064), topologically associated domains-TADs (3376 and 2031), and varying numbers of structural variations (SVs) in diploid and polyploid pitaya species, respectively. Overall, TAD boundaries were enriched with the motifs of AP2, WRKY18/60/75, MYB63/116, PHL2, and GATA8 in both pitaya species. By linking the open chromatin genomic structure to function, we identified the major changes in betalains biosynthesis pathway in diploid and polyploid pitaya. Moreover, the higher genetic expression of SmeADH1 [Chr11, Compartment A (135400000 - 135500000), genes inside the TAD region (135480000 - 135520000)], and lower expression of HuDOPA [Chr11, Compartment A (87100000 - 87200000), genes inside the TAD region (87160000-87200000)] acts as a key regulator of yellow and red color on the pericarp of polyploid and diploid pitaya, respectively. In addition, higher expression of HuCYP76AD1 genes in diploid pitaya and lower expression of SmeCYP76AD1 in polyploid pitaya potentially created the difference in the oxidase process that led to the production of betacyanin and betaxanthin, respectively. Furthermore, our results revealed not only the type of motifs that play a potential role in trait patterning but we also further uncovered that motif count in TAD-boundaries may impact the gene expression within the TAD regions of diploid and polyploid pitaya. Our valuable genomic resource and comparison of 3D euchromatin architecture of diploid and polyploid pitaya species will not only aid in the advancement of molecular breeding efforts but also offer insights into the organization of genomes, SVs, compartmentalization (A and B), and TADs, which have the potential to strengthen the idea of TADs-based trait improvement to achieve global food security.


