Maize (Zea mays L.) interaction with the arbuscular mycorrhizal fungus Rhizophagus irregularis allows mitigation of nitrogen deficiency stress: physiological and molecular characterization
Maize (Zea mays L.) interaction with the arbuscular mycorrhizal fungus Rhizophagus irregularis allows mitigation of nitrogen deficiency stress: physiological and molecular characterization
Decouard, B.; Chowdhury, N. B.; Saou, A.; Rigault, M.; Quillere, I.; Sapir, T.; Marmagne, A.; Paysant le Roux, C.; Launay-Avon, A.; Guerard, F.; Gakiere, B.; Mauve, C.; Levy-Leduc, C.; Barbillon, P.; Saha, R.; Hirel, B.; Courty, P.-E.; Wipf, D.; DELLAGI, A.
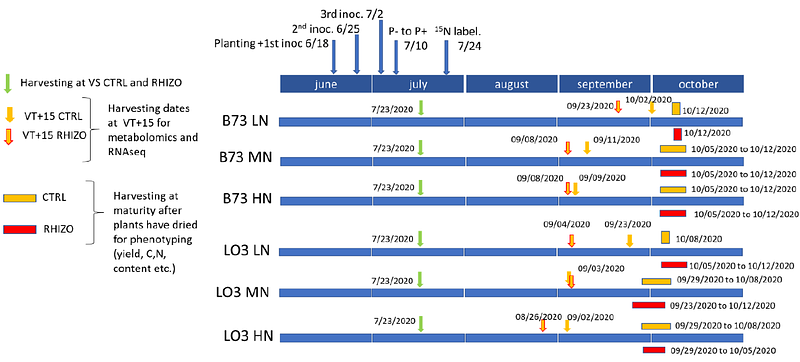
AbstractMaize is currently the most productive cereal crop in the world (www.faostat.org). It can associate with the Arbuscular Mycorrhizal Fungus (AMF) Rhizophagus irregularis which can provide additional water and mineral nutrients to the plant in return for C delivered by the host plant. Two maize lines were characterized at the physiological and molecular levels as they displayed contrasting responses to inoculation with the AMF when grown under optimal, medium, or low N fertilization conditions. For both these lines, the presence of the AMF allows development of a beneficial symbiotic association but only under limiting N fertilization conditions and allows to maintain plant biomass production when there is a five fold reduction in N supply. Physio-agronomical phenotyping and transcriptomic and metabolomic characterization of these two lines indicates that, (i) the level of N supply is the major factor affecting all studied traits; (ii) although the two lines display different transcriptomic and metabolomic responses to R. irregularis the agro-physiological traits are similar; and (iii) inoculation with the AMF relieves N deficiency stress. Analysis of the fungal transcriptome indicated that, like the plant transcriptome, it was also mainly affected by N nutrition level rather than by the maize host genotype. To explore potential metabolic pathways affected by the symbiosis, we integrated the transcriptomic data to model mycorrhized maize metabolism in a multi-organ Genome-scale metabolic model (GSM) iZMA6517, based on a stoichiometric approach. Both maize transcriptomics data integrated iZMA6517 model and R. irregularis transcriptome pointed to nucleotide and ureides metabolism as previously unexplored new drivers of the symbiotic N nutrition triggered by R. irregularis that allow maize growth improvement.