Patterns and drivers of genome-wide codon usage bias in the fungal order Sordariales
Patterns and drivers of genome-wide codon usage bias in the fungal order Sordariales
Hensen, N.; Hiltunen Thoren, M.; Johannesson, H.
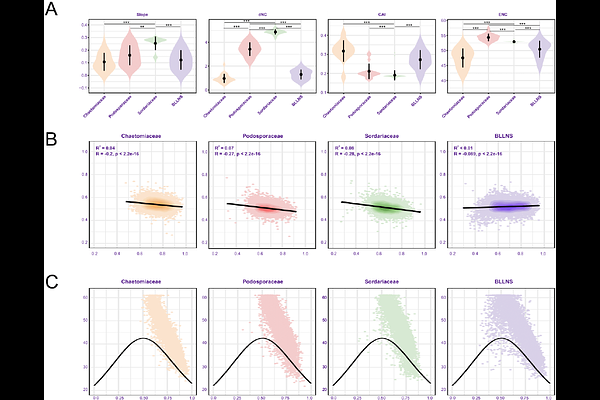
AbstractHere we present a study on amino acid composition, codon usage bias (CUB), and levels of selection driving codon usage in Sordariales fungi. We found that GC ending codons are used more often than AT ending codons in all Sordariales genomes, but the strength of CUB differs amongst families. The families Podosporaceae and Sordariaceae contain relatively low genome-wide levels of CUB, while the highest levels of CUB are found in Chaetomiaceae and the \"BLLNS\"-group, a monophyletic group of five other Sordariales families. Based on genomic clustering, and ancestral state reconstruction of GC nucleotides at the third codon position, we hypothesize that Podosporaceae and Sordariaceae represent the ancestral state of amino acid composition and CUB. The Chaetomiaceae and BLLNS have most likely diverged from this state, with increased natural selection driving use of specific codons, resulting in higher genome-wide CUB. We expect that the higher levels of CUB in Chaetomiaceae genomes might have been caused by ecological niche specialization, including high optimal growth temperature of some Chaetomiaceae species.