Genome-wide Mapping of Topoisomerase Binding Sites Suggests Topoisomerase 3α(TOP3A) as a Reader of Transcription-Replication Conflicts (TRC)
Genome-wide Mapping of Topoisomerase Binding Sites Suggests Topoisomerase 3α(TOP3A) as a Reader of Transcription-Replication Conflicts (TRC)
Zhang, H.; Sun, Y.; Saha, S.; Saha, L. K.; Pongor, L. S.; Dhall, A.; Pommier, Y.
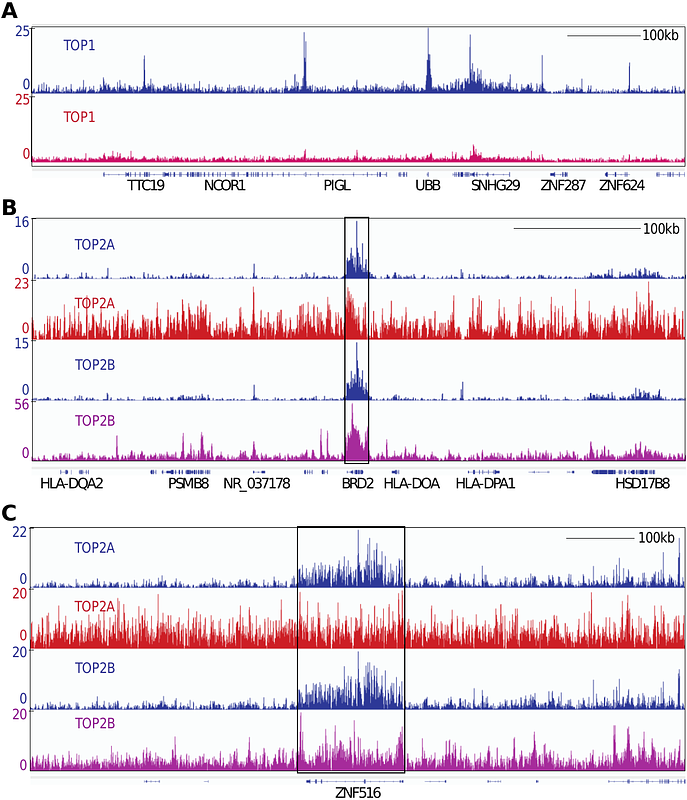
AbstractBoth transcription and replication can take place simultaneously on the same DNA template, potentially leading to transcription-replication conflicts (TRCs) and topological problems. Here we asked which topoisomerase(s) is/are the best candidate(s) for sensing TRC. Genome-wide topoisomerase binding sites were mapped in parallel for all the nuclear topoisomerases (TOP1, TOP2A, TOP2B, TOP3A and TOP3B). To increase the signal to noise ratio (SNR), we used ectopic expression of those topoisomerases in H293 cells followed by a modified CUT&Tag method. Although each topoisomerase showed distinct binding patterns, all topoisomerase binding signals positively correlated with gene transcription. TOP3A binding signals were suppressed by DNA replication inhibition. This was also observed but to a lesser extent for TOP2A and TOP2B. Hence, we propose the involvement of TOP3A in sensing both head-on TRCs (HO-TRCs) and codirectional TRCs (CD-TRCs). In which case, the TOP3A signals appear concentrated within the promoters and first 20 kb regions of the 5[prime] -end of genes, suggesting the prevalence of TRCs and the recruitment of TOP3A in the 5[prime]-regions of transcribed and replicated genes.


