Tracing of streptococcal strains from infant stool across human body sites links gut-specificity to adhesins
Tracing of streptococcal strains from infant stool across human body sites links gut-specificity to adhesins
Ormaasen, I.; Kjos, M.; Simpson, M. R.; Oien, T.; Snipen, L.; Rudi, K.
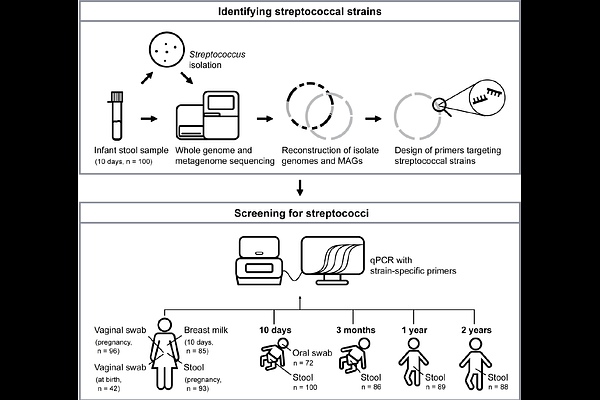
AbstractStreptococcal species are human commensals known to colonize multiple body sites. Despite being early gut colonizers, we lack strain-level information about their origin and persistence in the gut. To gain more insight into the habitats of the streptococci present in the infant gut, we did a systematic study where mother-infant pairs were sampled from multiple body sites (stool, oral cavity, vagina, breast milk). We performed whole metagenome sequencing and isolated streptococci from 100 infant stool samples (collected at 10 days of age). To trace the streptococci at the strain level, we designed selective qPCR primers for seven streptococcal strains, and these were later utilized to screen corresponding samples from multiple body sites of the infants and their mothers. We found that two of the strains (one Streptococcus parasanguinis and one Streptococcus vestibularis) were highly prevalent in stool samples, both from infants during their first 2 years of life and from their mothers, indicating that these strains are adapted to the gut environment. Interestingly, another S. parasanguinis strain, closely related to the gut-prevalent strain, showed a completely different prevalence pattern, and was mainly detected in vaginal swabs, breast milk and oral swabs. Comparisons of their genomes revealed major differences in genes encoding adhesins, suggesting that host surface attachment could be a key factor for the observed differences in body site specificity. Together, our extensive tracing of streptococci across body sites of 100 infants and their mothers, provide strain-level information of prevalence patterns and reveal the presence of gut-specific streptococci.