Patterning defects in mice with defective ventricular wall maturation and cardiomyopathy
Patterning defects in mice with defective ventricular wall maturation and cardiomyopathy
Santos-cantador, J.; Siguero-Alvarez, M.; de la Pompa, J. L.
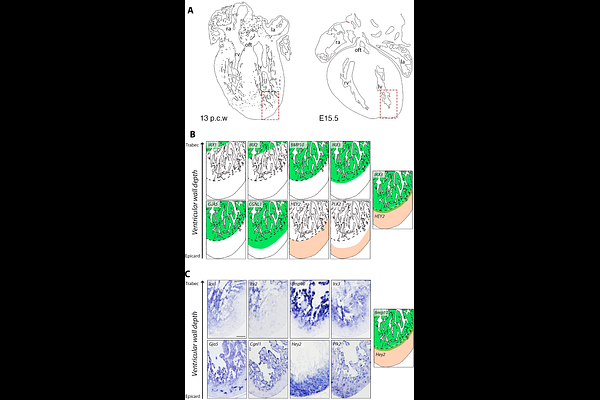
AbstractVentricular chamber development involves the coordinated maturation of diverse cell populations. In the human fetal heart, single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics reveal marked regional gene expression differences. In contrast, the mouse ventricular wall appears more homogeneous, except for a transient hybrid cardiomyocyte population co-expressing compact (Hey2) and trabecular (Irx3, Nppa, Bmp10) markers, indicating a transitional lineage state. To further investigate this, we used in situ hybridization (ISH) to examine the expression of a selected set of markers in normal and left ventricular non-compaction cardiomyopathy (LVNC) mouse models. In developing mouse ventricles, the expression of key marker genes was largely restricted to two wide myocardial domains, compact and trabecular myocardium, suggesting a less complex regional organization than human fetal heart. Other markers labelled endocardial and coronary endothelial cells rather than cardiomyocytes, differing from patterns observed in the human heart. In the LVNC model, various markers exhibited altered spatial expression, indicating that precise regional organization of gene expression is critical for normal ventricular wall maturation. These findings underscore the critical role of spatially regulated gene programs in ventricular chamber development and point to their potential involvement in cardiomyopathy pathogenesis.