High-resolution chromosome-level genome provides molecular insights into adaptive evolution in crabs
High-resolution chromosome-level genome provides molecular insights into adaptive evolution in crabs
Zhang, Y.; Yuan, Y.; Zhang, M.; Yu, X.; Qiu, B.; Wu, F.; Tocher, D. R.; Zhang, J.; Ye, S.; Cui, W.; Leung, J. Y. S.; Ikhwanuddin, M.; Waqas, W.; Dildar, T.; Ma, H.
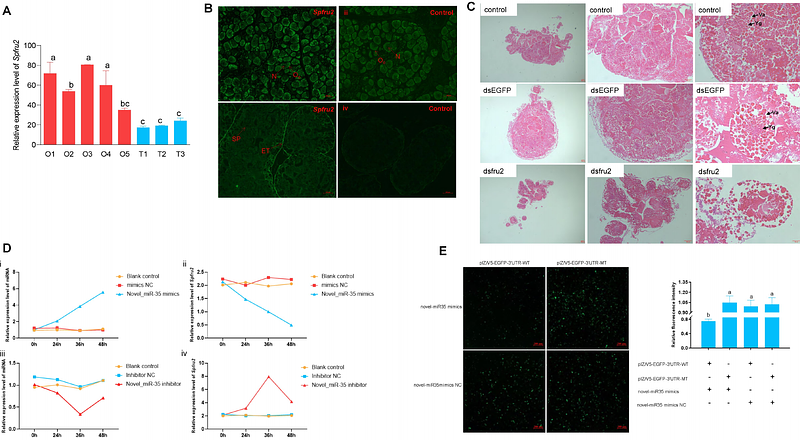
AbstractCrabs thrive in diverse ecosystems, from coral reefs to hydrothermal vents and terrestrial habitats. Here, we report a comprehensive genomic analysis of the mud crab using ultralong sequencing technologies, achieving a high-quality chromosome-level assembly. The refined 1.21 Gb genome, with an impressive contig N50 of 11.45 Mb, offers a valuable genomic resource. Gene family analysis shows expansion in development-related pathways and contraction in metabolic pathways, indicating niche adaptations. Notably, Investigation into Hox gene regulation sheds light on their role in pleopod development, with the Abd-A gene identified as a linchpin. Posttranscriptional regulation involving novel-miR1317 negatively regulates Abd-A levels. Furthermore, the fru gene\'s potential role in ovarian development and the identification of novel-miRNA-35 as a regulator of Spfru2 add complexity to gene regulatory networks. Comparative functional analysis across Decapoda species reveals neofunctionalization of the elovl6 gene in the synthesis of long-chain polyunsaturated fatty acids (LC-PUFA), suggesting its importance in environmental adaptation. These findings contribute significantly to our understanding of crab adaptability and evolutionary dynamics, offering a robust foundation for future investigations.


