Beyond binding motif: genomic context predicts transcription factor dependent regulation in Arabidopsis
Beyond binding motif: genomic context predicts transcription factor dependent regulation in Arabidopsis
Turchi, L.; Lucas, J.; Tichtinsky, G.; Thierry-Mieg, N.; Blanc-Mathieu, R.; Parcy, F.; Frenoy, A.
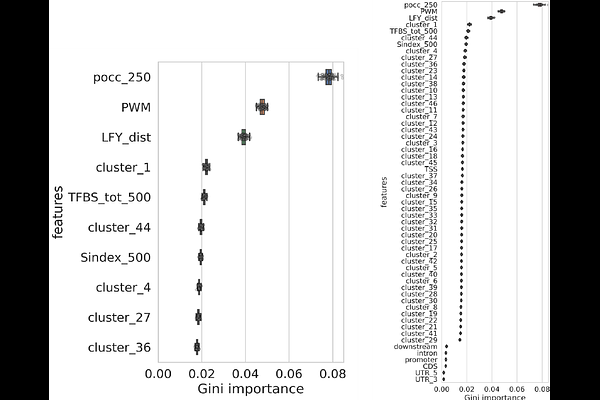
AbstractTranscription Factors (TFs) play a crucial role in the spatiotemporal control of gene expression. Despite the presence of many potential TF binding sites (TFBS) across the genome, TFs do not interact with all of them, and, among those, only a subset leads to actual regulation. In this work, we investigate how the genomic context of a putative TFBS, including the occurrence of binding motifs for other TFs, can be used to predict effective transcriptional regulation by a TF of interest. We focus on LEAFY (LFY), a plant-specific TF and master regulator of flower development. Using available transcriptomes and TF-DNA binding experiments, we identify 1164 LFY binding sites associated with a regulatory response in Arabidopsis thaliana. We then apply a machine learning approach based on properties of the surrounding genomic region, to discriminate these regulatory LFY binding sites from non-regulatory ones. Detailed analysis of the model\'s components reveals that, for LFY, the density and quality of binding sites constitute the most important features but were not sufficient on their own to predict regulatory activity. The presence of binding sites for other transcription factors and the overall richness in TFBS were also essential. These results clarify the nature of the regulatory code by which LFY operates and could serve as a basis for the study of the regulatory elements of other transcription factors.