Fundamental limits on patterning by Turing-like reaction-diffusion mechanisms
Fundamental limits on patterning by Turing-like reaction-diffusion mechanisms
Muzatko, D.; Daga, B.; Hiscock, T. W.
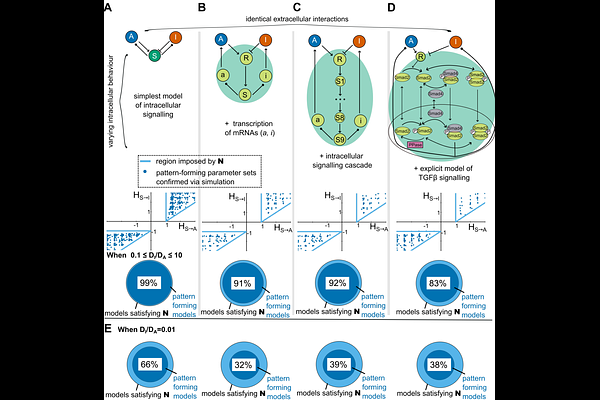
AbstractTuring\'s longstanding reaction-diffusion hypothesis explains how molecular patterns can self-organize de novo in otherwise homogeneous tissues. However, whilst simple Turing models can qualitatively recapitulate patterning in silico, they are often highly simplified approximations of the molecular complexity operating in vivo. Here, we investigate significantly more complex reaction-diffusion models that seek to directly capture the mechanisms involved in intercellular signalling. Rather than resulting in minor, quantitative differences, we find that these more realistic systems display qualitatively distinct behaviours compared to simple Turing models. By combining large-scale simulations with formal mathematical proofs, we show, rather generally, that patterning is strongly constrained by the extracellular interactions in the system but is relatively insensitive to the intracellular dynamics assumed. When applied to the activator-inhibitor paradigm, we find a broader repertoire of self-organizing circuits than previously recognized, including some which are unexpectedly robust to parameters. Beyond these examples, we have packaged our highly performant numerical methods into a freely available and easy-to-use software pipeline, ReactionDiffusion.jl, that allows arbitrarily complex reaction-diffusion systems to be simulated at scale.