Single-oocyte transcriptional profile of early-stage human oocytes reveals differentially expressed genes in the primordial and transitioning stages.
Single-oocyte transcriptional profile of early-stage human oocytes reveals differentially expressed genes in the primordial and transitioning stages.
Machlin, J. H.; Hannum, D. F.; Jones, A. S.; Schissel, T.; Potosky, K.; Marsh, E.; Hammoud, S.; Padmanabhan, V.; Li, J.; Shikanov, A.
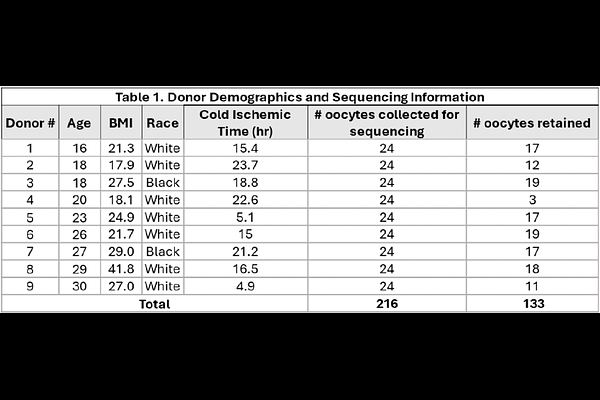
AbstractThe critical initial step in human oocyte maturation - the transition of ovarian follicles from dormancy to activation - remains poorly understood. Here we performed RNA sequencing on single oocytes isolated from early-stage follicles from nine healthy reproductive-age donors. Data for 133 high-quality oocytes formed two connected clusters, C1 and C2, with 5,449 significantly differentially expressed genes. Using recently reported gene lists for early-stage follicles we found that C1 oocytes likely came from earlier, dormant primordial follicles, while C2 oocytes match later-stage primordial or transitioning follicles. We sought to validate two DE genes, UHRF1 for C1 and CCN2 for C2, by their RNA-FISH in situ pattern in morphologically classified follicles, but did not observe statistically significant differences between follicle stages. This apparent discrepancy between the stage of the follicle determined by its morphology and the transcriptional state of the oocyte, if replicated in additional studies, may indicate a lack of closely coupled synchrony between follicle morphology and the functional state of the oocyte.