Cell Morphology and Biophysical Mechanisms-Informed Traction Force Microscopy Using Machine Learning
Cell Morphology and Biophysical Mechanisms-Informed Traction Force Microscopy Using Machine Learning
Fujiwara, K.; Fujikawa, R.; Suzuki, Y.; Suzuki, K. T.; Sakumura, Y.
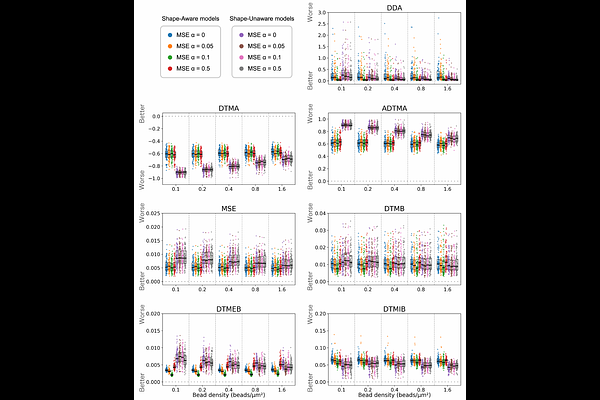
AbstractQuantitative evaluation of cell motility is essential for understanding biological functions. Traction force microscopy (TFM) is a method for quantifying cellular traction forces. By performing inverse mathematical analysis of substrate deformations induced by cellular forces, it is possible to estimate the underlying traction forces. Among various approaches for detecting substrate deformations, the most widely used is to track the displacement of fluorescent beads randomly embedded in the substrate. However, it is well known that the accuracy of force estimation deteriorates when the observation density is low. Furthermore, standardized datasets have not been established, as validation settings-such as random force distributions on the substrate, cell size, and bead density-vary across studies. To enable quantitative evaluation, we constructed a dataset based on the biophysical mechanism by which cellular traction forces are transmitted to the substrate through stress fibers, clutch proteins, and focal adhesions. In addition, we proposed a novel machine learning model that incorporates cell shape information obtained from observations, and we designed a loss function that accounts for both force magnitude and direction. As a result, our method enables highly accurate force estimation even under sparse observation conditions.