Controlling semiconductor growth with structured de novo protein interfaces
Controlling semiconductor growth with structured de novo protein interfaces
Saragovi, A.; Pyles, H.; Kwon, P.; Hanikel, N.; Davila-Hernandez, F. A.; Bera, A.; Kang, A.; Brackenbrough, E.; Vafeados, D. K.; Allen, A.; Stewart, L.; Baker, D.
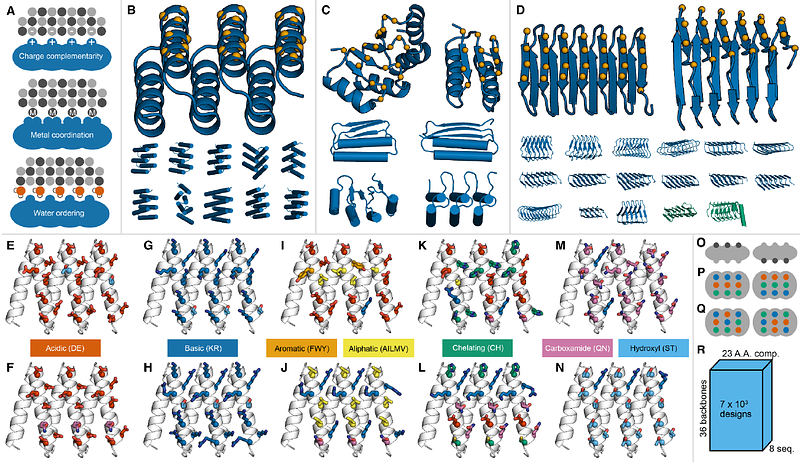
AbstractProtein design now enables the precise arrangement of atoms on the length scales (nanometers) of inorganic crystal nuclei, opening up the possibility of templating semiconductor growth. We designed proteins presenting regularly repeating interfaces presenting functional groups likely to organize ions and water molecules, and characterized their ability to bind to and promote nucleation of ZnO. Utilizing the scattering properties of ZnO nanoparticles, we developed a flow cytometry based sorting methodology and identified thirteen proteins with ZnO binding interfaces. Three designs promoted ZnO nucleation under conditions where traditional inorganic binding peptides and control proteins were ineffective. Incorporation of these interfaces into higher order assemblies further enhanced nucleation. These findings demonstrate the potential of using protein design to modulate semiconductor growth and generate protein-semiconductor hybrid materials.


