Pan-genome analysis of different morphotypes reveals genomic basis of Brassica oleracea domestication and differential organogenesis
Pan-genome analysis of different morphotypes reveals genomic basis of Brassica oleracea domestication and differential organogenesis
Guo, N.; Wang, S.; Wang, T.; Duan, M.; Zong, M.; Miao, L.; Han, S.; Wang, G.; Liu, X.; Zhang, D.; Jiao, C.; Xu, H.; Chen, L.; Fei, Z.; Li, J.; Liu, F.
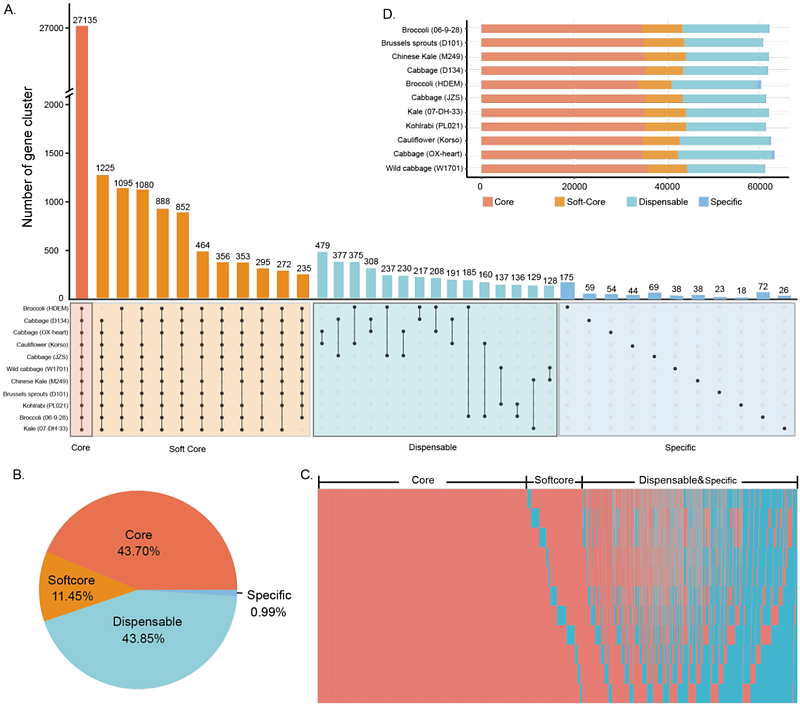
AbstractThe domestication of Brassica oleracea has resulted in diverse morphological types with distinct patterns of organ development. Here we report a graph-based pan-genome of B. oleracea constructed with high-quality genome assemblies of different morphotypes. The pan-genome harbors over 200 structural variant (SV) hotspot regions enriched with auxin and flowering-related genes. Population genomic analyses reveal that early domestication of B. oleracea focused on leaf or stem selection. Gene flows resulting from agricultural practices and variety improvement are detected among different morphotypes. Selective sweep analysis identifies an auxin-responsive SAUR gene and a CLE family gene as the crucial players in the leaf-stem differentiation during the early stage of B. oleracea domestication, and the BoKAN1 gene as instrumental in shaping the leafy heads of cabbage and Brussels sprouts. Our pan-genome and functional analyses further discover that variations in the BoFLC2 gene play key roles in the divergence of vernalization and flowering characteristics among different morphotypes, and variations in the first intron of BoFLC3 are involved in fine-tuning the flowering process in cauliflower. This study provides a comprehensive understanding of the pan-genome of B. oleracea and sheds light on the domestication and differential organ development of this globally important crop species.