TICKHUNTER: A Targeted Hybridization-Capture Sequencing Approach for the Detection and Characterization of Tick-borne Pathogens and Blood Meals
TICKHUNTER: A Targeted Hybridization-Capture Sequencing Approach for the Detection and Characterization of Tick-borne Pathogens and Blood Meals
Wang, X.; Brown, J. J.; Lilly, M. V.; Plimpton, L.; Murrell, C. M.; Llanos-Soto, S. G.; Zehr, J. D.; Sun, Y.; Reboul, G.; Sams, K.; Sabaawy, A.; Singh, L.; Tallmadge, R. L.; Mitchell, P. K.; Anderson, R. R.; Ladd-Wilson, S. G.; Bourgikos, E.; Vogels, C. B.; Olarte-Castillo, X. A.; Diuk-Wasser, M.; Goodman, L. B.
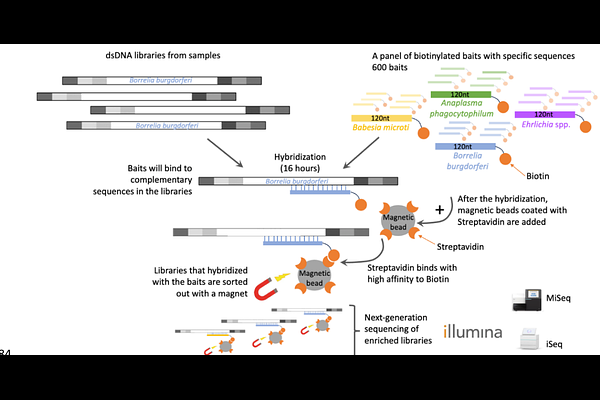
AbstractTicks are vectors of many debilitating pathogens that are transmitted during blood meal acquisition. As both vector and pathogen species continue to emerge, there is a critical need for improved pathogen diagnostic and host remnant identification methods to develop interventions. Current pathogen detection methods are limited in their ability to subtype and detect emerging variants and non-vector borne \'look-alike\' diseases. Determination of tick blood meal sources is also an elusive step for understanding tick interactions with the biotic community in a given environment, with consequences for environmental management and disease prevention strategies. We have created an accessible hybridization capture next-generation sequencing panel called TICKHUNTER to improve molecular detection and subtyping of bacterial and parasitic pathogens in both clinical and tick samples. For disease ecology studies, the panel also detects host blood meal sources of interest. An alternative method for unbiased blood meal remnant analysis is also presented. We find that TICKHUNTER is a promising tool for effective and accurate detection of a variety of tick-borne pathogens, based on comparable linearity, limit of detection, sensitivity, and specificity to real-time PCR. Additionally, it can detect and characterize unexpected pathogens due to the large capacity for multiplexing and flexibility in variant detection.