Mycobacterium tuberculosis Ku stimulates multi-round DNA unwinding by UvrD1 monomers
Mycobacterium tuberculosis Ku stimulates multi-round DNA unwinding by UvrD1 monomers
Chadda, A.; Kozlov, A. G.; Nguyen, B.; Lohman, T. M.; Galburt, E.
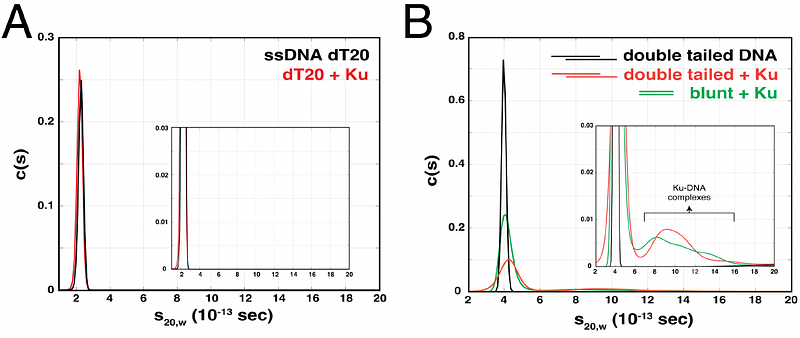
AbstractMycobacterium tuberculosis is the causative agent of Tuberculosis. During the host response to infection, the bacterium is exposed to both reactive oxygen species and nitrogen intermediates that can cause DNA damage. It is becoming clear that the DNA damage response in Mtb and related actinobacteria function via distinct pathways as compared to well-studied model bacteria. For example, we have previously shown that the DNA repair helicase UvrD1 is activated for processive unwinding via redox-dependent dimerization. In addition, mycobacteria contain a homo-dimeric Ku protein, homologous to the eukaryotic Ku70/Ku80 dimer, that plays roles in double-stranded break repair via non-homologous end-joining. Ku has been shown to stimulate the helicase activity of UvrD1, but the molecular mechanism, as well as which redox form of UvrD1 is activated, is unknown. We show here that Ku specifically stimulates multi-round unwinding by UvrD1 monomers which are able to slowly unwind DNA, but at rates 100-fold slower than the dimer. We also demonstrate that the UvrD1 C-terminal Tudor domain is required for the formation of a Ku-UvrD1 protein complex and activation. We show that Mtb Ku dimers bind with high nearest neighbor cooperativity to duplex DNA and that UvrD1 activation is observed when the DNA substrate is bound with two or three Ku dimers. Our observations reveal aspects of the interactions between DNA, Mtb Ku, and UvrD1 and highlight the potential role of UvrD1 in multiple DNA repair pathways through different mechanisms of activation.