Comprehensive detection of structural variations in long and short reads dataset of French cattle
Comprehensive detection of structural variations in long and short reads dataset of French cattle
Naji, M. M.; Klopp, C.; Faraut, T.; Eche, C.; Di Franco, A.; Birbes, C.; Marcuzzo, C.; Suin, A.; Iampietro, C.; Kuchly, C.; Vernette, C.; Fritz, S.; Grohs, C.; Gaspin, C.; Milan, D.; Donnadieu, C.; Boichard, D.; Sanchez, M.-P.; Boussaha, M.
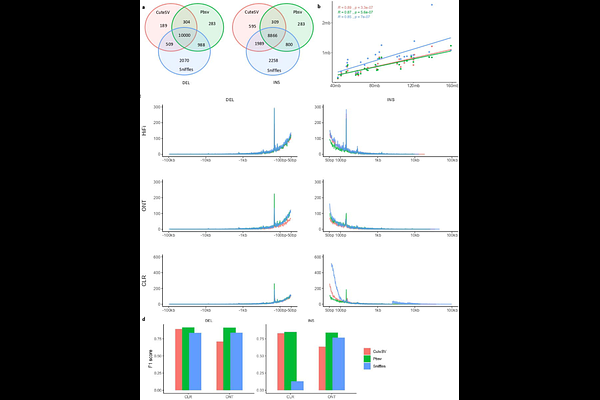
AbstractStructural variants (SVs) correspond to different types of genomic variants larger than 50 bp. Many findings suggest the use of long rather than short reads to improve the accuracy of SV detection. Here, we present the results of an in-depth analysis for detection of SVs, mainly large insertions and deletions, in 14 French bovine breeds, based on whole-genome data comprising 176 long-read and 571 short-read samples, with 154 individuals having both long- and short-read data available. We first investigated possible biases on the performances of well-known SV detection tools, namely CUTESV, PBSV, and SNIFFLES, using long reads from different technologies, including PacBio HiFi, Oxford ONT, and PacBio CLR. We subsequently highlighted the abilities of tools for detecting SVs (DELLY, LUMPY, and MANTA) and for genotyping known SVs (GRAPHTYPER, SVTYPER, PARAGRAPH, and VG toolkit) using short-read data. We then show how the incremental composition of samples in the reference panel affected the SV genotyping for six validation individuals sequenced in short reads. We then searched for the optimal parameters and created the final SV reference panel consisting of 25,191 deletions and 30,118 insertions. Finally, we emphasized the landscape of the genotyped SVs segregating across 571 short-read individuals of 14 breeds.