Genetic diversity in gene regulatory interactions underlies the response to soil drying in the model grass Brachypodium distachyon
Genetic diversity in gene regulatory interactions underlies the response to soil drying in the model grass Brachypodium distachyon
Yun, J.; Burnett, A. C.; Rogers, A.; Des Marais, D. L.
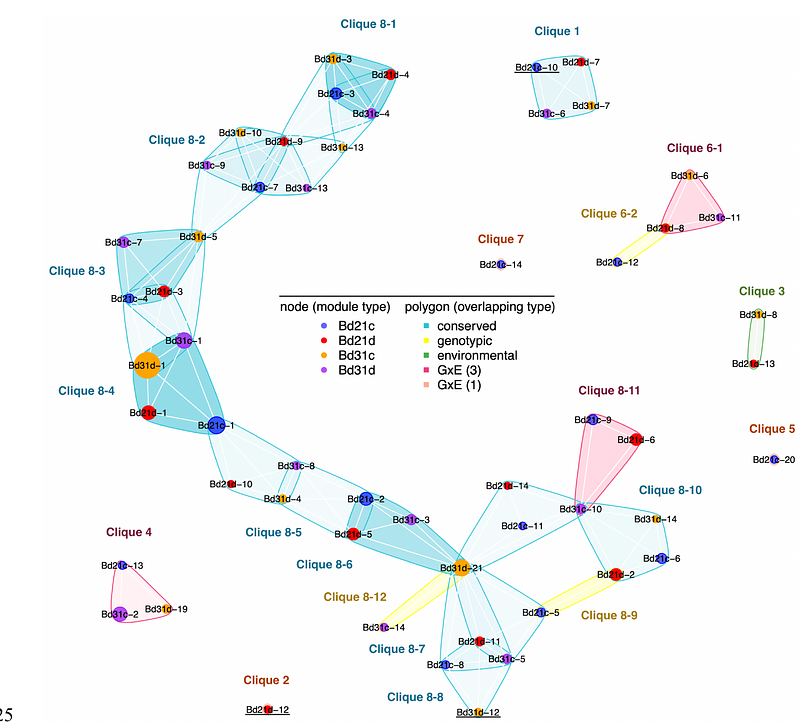
AbstractGene expression is a quantitative trait under the control of genetic and environmental factors and their interaction, so-called GxE. Understanding the mechanisms driving GxE is fundamental for ensuring stable crop performance across environments, and for predicting the response of natural populations to climate change. Gene expression is regulated through complex molecular networks, however environmental and genotypic effects on genome-wide regulatory networks are rarely considered. In this study, we model genome-scale gene expression variation between two natural accessions of the model grass Brachypodium distachyon and their response to soil drying. We identified genotypic, environmental, and GxE responses in physiological, metabolic, and gene expression traits. We then identified gene regulation conservation and variation among conditions and genotypes, simplified as co-expression clusters found unique in or conserved across library types. Putative gene regulatory interactions are inferred as network edges with a graphical modelling approach, resulting in hypotheses about gene-gene interactions specific to -- or with higher affinity in -- one genotype, one environmental treatment, or in one genotype under treatment. We further find that some gene-gene interactions are conserved across conditions such differential expression is apparently transmitted to the target gene. These variably detected edges cluster together in co-expression modules, suggestive of different constraints or selection strength acting on specific pathways. We further applied our graphical modeling approach to identify putative, environmentally dependent regulatory mechanisms of leaf glucose content as an exemplar metabolite. Our study highlights an approach to identify variable features of gene regulatory networks and thereby identify key components for later genomic intervention to elucidate function or modulate environmental response. Our results also suggest possible targets of evolutionary change in gene regulatory networks associated with environmental plasticity.


