Disentangling abiotic and biotic effects of treated wastewater on stream biofilm resistomes enables the discovery of a new planctomycete beta-lactamase
Disentangling abiotic and biotic effects of treated wastewater on stream biofilm resistomes enables the discovery of a new planctomycete beta-lactamase
Attrah, M.; Schaerer, M. R.; Esposito, M.; Gionchetta, G.; Buergmann, H.; Lens, P.; Fenner, K.; van de Vossenberg, J.; Robinson, S. L.
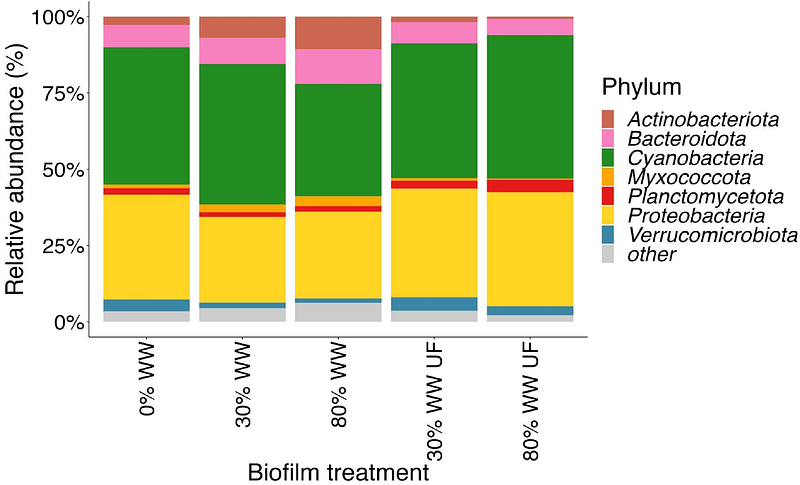
AbstractAntibiotic resistance, mediated by environmental reservoirs, poses a threat to human and animal health. Aquatic biofilms impacted by treated wastewater (WW) are known environmental reservoirs for antibiotic resistance, but it remains unclear if primarily biotic or abiotic factors from WW influence the abundance and dissemination of antibiotic resistance genes within biofilm communities. To disentangle these factors, natural biofilms were previously grown in flume systems with different proportions of stream water and either ultrafiltered or non-ultrafiltered treated WW. In this study, we conducted deep shotgun metagenomic sequencing of 75 biofilm, stream, and WW samples from these flume systems and compared microbiome and resistome composition. Our analysis revealed a strong relationship between the resistome and prokaryotic microbiome composition which was also associated with experimental treatment. Within biofilm communities exposed to increasing percentages of WW, we observed a decrease in the relative abundances of Cyanobacteriota and Verrucomicrobiota, while Bacteroidota increased. Notably, several antibiotic resistance genes (ARGs) exhibited a significant increase in abundance in biofilms grown with increasing WW%, while a small number of sulfonamide and beta-lactamase ARGs showed higher abundance when biofilms were instead grown in ultrafiltered WW. Overall, our results pointed toward the dominance of biotic over abiotic factors in determining ARG abundances in WW-impacted stream biofilms. However, some ARGs exhibited divergent abundance patterns through proposed gene family-specific mechanisms. To investigate these gene families experimentally, we biochemically characterized a new, sequence-divergent beta-lactamase from planctomycetes abundant in stream biofilms and in WW. This ARG discovery is noteworthy, not only due to its unusual source organism belonging to the phylum Planctomycetota, but also because it was not detected by standard computational ARG tools. This underpins the need for continued development of ARG detection methods using workflows with in silico sequence and structure analysis coupled to in vitro functional characterization. In summary, this study assessed the relative importance of biotic and abiotic factors in the context of WW discharge and its impact on antibiotic resistance genes in aquatic ecosystems.