Genome graphs reveal the importance of structural variation in Mycobacterium tuberculosis evolution and drug resistance
Genome graphs reveal the importance of structural variation in Mycobacterium tuberculosis evolution and drug resistance
Canalda-Baltrons, A.; Silcocks, M.; Hall, M. B.; Theys, D.; Chang, X.; Viberg, L. T.; Sherry, N. L.; Coin, L.; Dunstan, S. J.
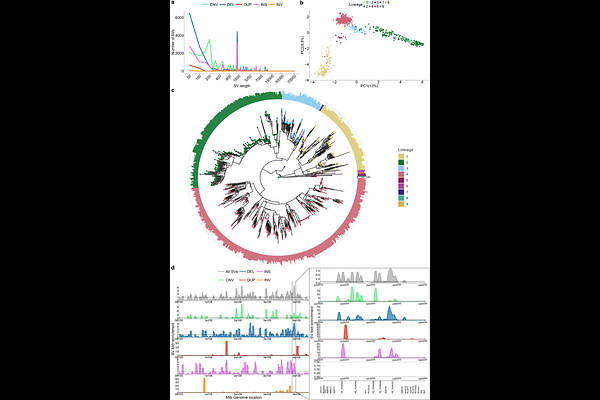
AbstractStructural variants (SVs) are increasingly recognized as key drivers of bacterial evolution, yet their role has not been explored thoroughly. This is due to limitations in traditional short-read sequencing and linear reference-based analyses, which can miss complex structural changes. Tuberculosis (TB), a disease caused by Mycobacterium tuberculosis (Mtb), remains a major global health concern. In this study, we harness long-read sequencing technologies and genome graph tools to construct a Mtb pangenome reference graph (PRG) from 859 high-quality, diverse, long-read assemblies. To enable accurate genotyping of SVs leveraging the PRG, we developed miniwalk, a tool that outperforms a traditional linear genome-based approach in precision for SV detection. We characterize patterns of structural variation genome-wide, revealing a virulence-associated ESX-5 deletion to be recurrent across the phylogeny, and fixed in a sub-lineage of L4. Systematic screens for additional genes that are recurrently affected by SVs implicated those related to metal homeostasis, including a copper exporter fixed in the widely distributed L1.2.1 sub-lineage. Lastly, we genotyped 41,134 isolates and found SVs putatively associated with resistance to various first and second-line drugs. These findings underscore the broader role of SVs in shaping Mtb diversity, highlighting their importance in both understanding evolution and designing strategies to combat drug-resistant TB.